2BYU
 
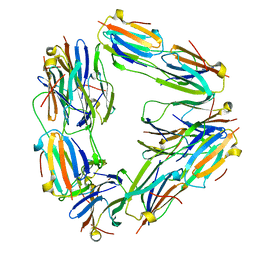 | | Negative stain EM reconstruction of M.tuberculosis Acr1(Hsp 16.3) fitted with wheat sHSP dimer | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Kennaway, C.K, Benesch, J.L.P, Gohlke, U, Wang, L, Robinson, C.V, Orlova, E.V, Saibil, H.R, Keep, N.H. | | Deposit date: | 2005-08-05 | | Release date: | 2005-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dodecameric Structure of the Small Heat Shock Protein Acr1 from Mycobacterium Tuberculosis.
J.Biol.Chem., 280, 2005
|
|
1J4H
 
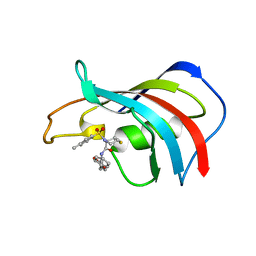 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
5W0Z
 
 | |
4V9D
 
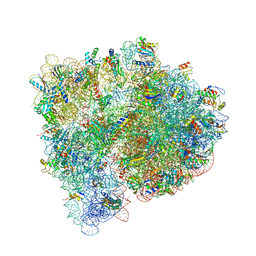 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
3BIY
 
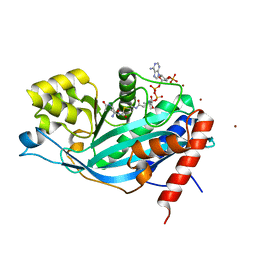 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
7OJT
 
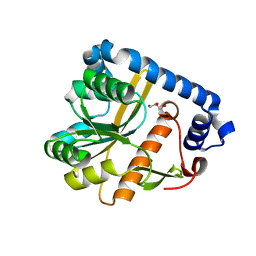 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
6NPY
 
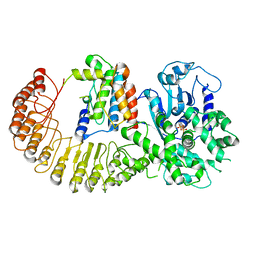 | | Cryo-EM structure of NLRP3 bound to NEK7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 3, ... | | Authors: | Sharif, H, Wang, L, Wang, W.L, Wu, H. | | Deposit date: | 2019-01-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome.
Nature, 570, 2019
|
|
5W1C
 
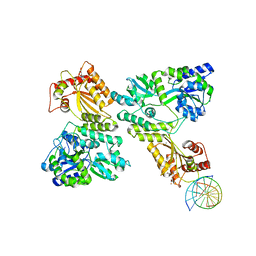 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W0R
 
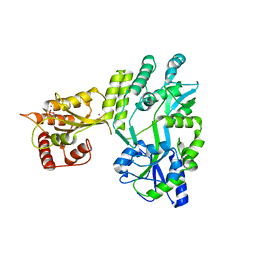 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | Descriptor: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W0U
 
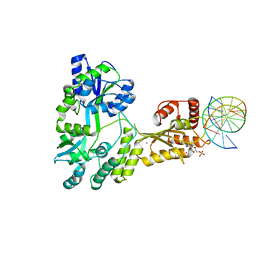 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
8I4B
 
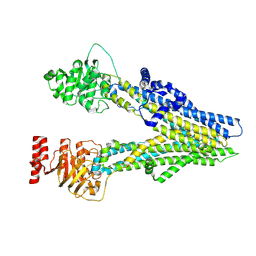 | | Cryo-EM structure of apo-form ABCC4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4A
 
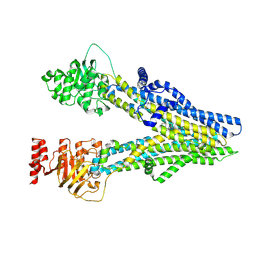 | | Cryo-EM structure of dipyridamole-bound ABCC4 | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4C
 
 | | Cryo-EM structure of U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8HP6
 
 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | Descriptor: | (S)-2-haloacid dehalogenase, SODIUM ION | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP5
 
 | | Crystal structure of (S)-2-haloacid dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP7
 
 | | Crystal structure of (S)-2-haloacid dehalogenase K152A mutant trapped with (2R)-4-amino-2-hydroxybutanoic acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
4WQN
 
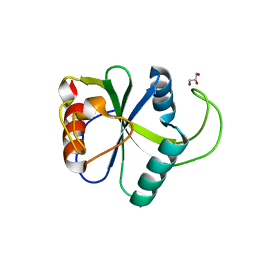 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
7BU1
 
 | |
7BTZ
 
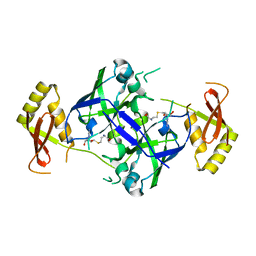 | | Crystal structure of TrmO | | Descriptor: | Putative tRNA (Adenine(37)-N6)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fan, C.P, Wang, L, Hu, W.H, Yang, C.W. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of TrmO from Pseudomonas aeruginosa
To Be Published
|
|
1CM0
 
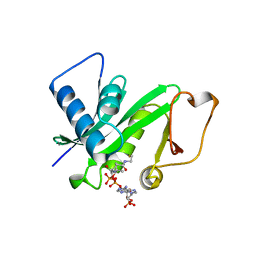 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
6JBJ
 
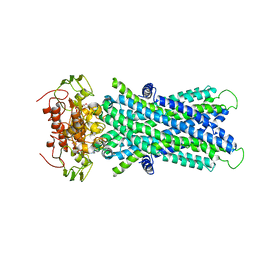 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
2VA1
 
 | | Crystal structure of UMP kinase from Ureaplasma parvum | | Descriptor: | PHOSPHATE ION, URIDYLATE KINASE | | Authors: | Egeblad-Welin, L, Welin, M, Wang, L, Eriksson, S. | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Investigations of Ureaplasma Parvum Ump Kinase - a Potential Antibacterial Drug Target
FEBS J., 274, 2007
|
|
1YD2
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y19F bound to the catalytic divalent cation | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
1YD6
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Bacillus caldotenax | | Descriptor: | CHLORIDE ION, SULFATE ION, UvrC | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
4R99
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
