1Y88
 
 | | Crystal Structure of Protein of Unknown Function AF1548 | | Descriptor: | CHLORIDE ION, Hypothetical protein AF1548, SULFATE ION | | Authors: | Lunin, V.V, Evdokimova, E, Kudritskaya, M, Cuff, M.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of hypothetical protein AF1548 from Archaeoglobus fulgidus
To be Published
|
|
1Y0N
 
 | | Structure of Protein of Unknown Function PA3463 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Hypothetical UPF0270 protein PA3463 | | Authors: | Binkowski, T.A, Edwards, A, Savchenko, A, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein PA3463 from Pseudomonas aeruginosa strain PAO1
To be Published
|
|
1Y0K
 
 | | Structure of Protein of Unknown Function PA4535 from Pseudomonas aeruginosa strain PAO1, Monooxygenase Superfamily | | Descriptor: | hypothetical protein PA4535 | | Authors: | Nocek, B.P, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 A Crystal Structure of the Hypothetical Protein Pa4535 from Pseudomonas Aeruginosa
To be Published
|
|
1Y7P
 
 | | 1.9 A Crystal Structure of a Protein of Unknown Function AF1403 from Archaeoglobus fulgidus, Probable Metabolic Regulator | | Descriptor: | Hypothetical protein AF1403, ZINC ION, beta-D-ribopyranose | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of a hypothetical protein AF1403 from Archaeoglobus fulgidus
To be Published
|
|
1KS2
 
 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
7ROA
 
 | | Crystal structure of EntV136 from Enterococcus faecalis | | Descriptor: | EntV | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-07-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
4XA9
 
 | | Crystal structure of the complex between the N-terminal domain of RavJ and LegL1 from Legionella pneumophila str. Philadelphia | | Descriptor: | Gala protein type 1, 3 or 4, Uncharacterized protein | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Sul3 | | Authors: | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2J
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 apoenzyme | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | Descriptor: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
4XI1
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, wild-type | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, GLYCEROL, ... | | Authors: | Stogios, P.J, Quaile, T, Skarina, T, Cuff, M, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4R5Q
 
 | | Crystal structure and nuclease activity of the CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis containing a [2Fe-2S] cluster | | Descriptor: | CRISPR-associated exonuclease, Cas4 family, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nocek, B, Skarina, T, Lemak, S, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure and nuclease activity of the CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis containing a [2Fe-2S] cluster
To be Published
|
|
4JHC
 
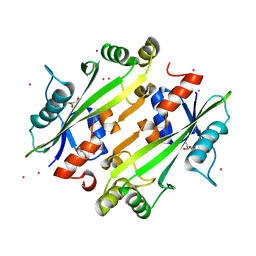 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli | | Descriptor: | GLYCEROL, Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2013-03-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
1WPB
 
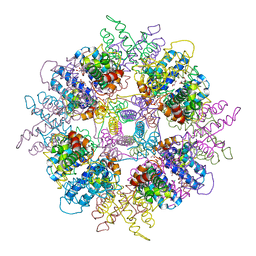 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
5FBT
 
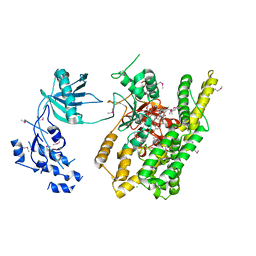 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin | | Descriptor: | CHLORIDE ION, Phosphoenolpyruvate synthase, Rifampin | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
1XEB
 
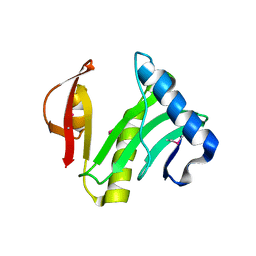 | | Crystal Structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA0115 | | Authors: | Bertero, M.G, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A.E, Savchenko, A, Strynadka, N, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa
To be Published
|
|
5FBS
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with ADP and magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoenolpyruvate synthase | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
6NLP
 
 | | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113 | | Descriptor: | 1,2-ETHANEDIOL, Bacterial extracellular solute-binding family protein, IMIDAZOLE | | Authors: | Tan, K, SKarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113
To Be Published
|
|
4WZ2
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | Authors: | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
3OKX
 
 | | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris | | Descriptor: | S-ADENOSYLMETHIONINE, YaeB-like protein RPA0152 | | Authors: | Chang, C, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris
To be Published
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
4WZ3
 
 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
6AOJ
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal yeast Hog1p sequence | | Descriptor: | CHLORIDE ION, Ceg4, MAGNESIUM ION | | Authors: | Stogios, P.J, Nocek, B, Cuff, M.E, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
