8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
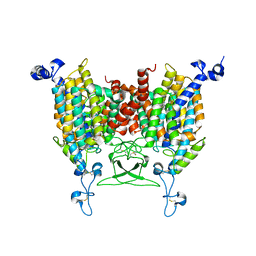 | | 6-BAP bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
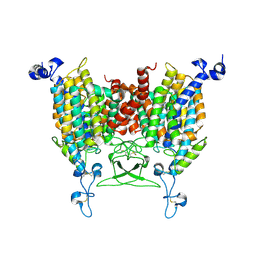 | | kinetin bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
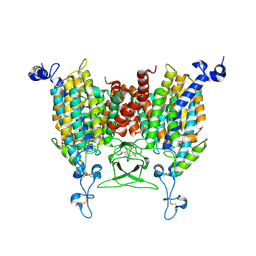 | | Endogenous substrate adenine bound state of Arabidopsis AZG1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRO
 
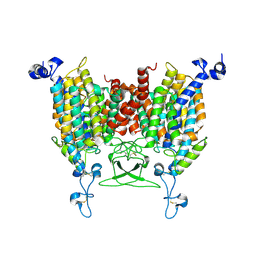 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
7XUK
 
 | |
7XUN
 
 | |
7KNX
 
 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNW
 
 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
8YJO
 
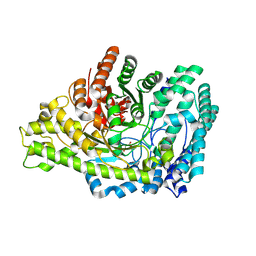 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJN
 
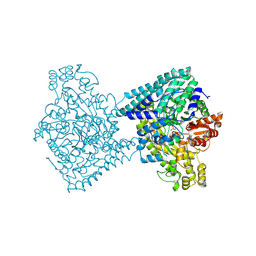 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8IOY
 
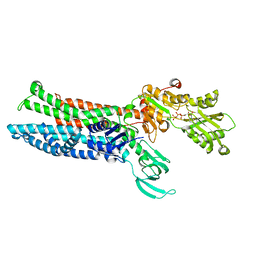 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
2P3T
 
 | |
1IHT
 
 | |
1IHS
 
 | |
2P3U
 
 | |
6M2W
 
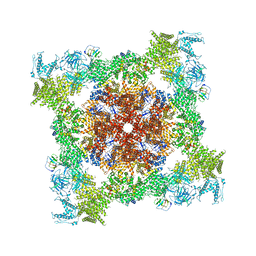 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
4WTR
 
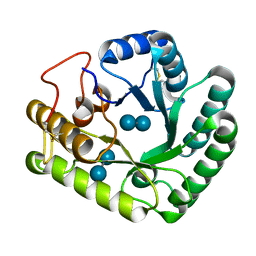 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTP
 
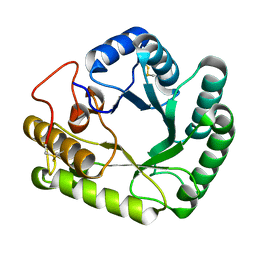 | | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei | | Descriptor: | 1,2-ETHANEDIOL, beta-1,3-glucanosyltransferase | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTS
 
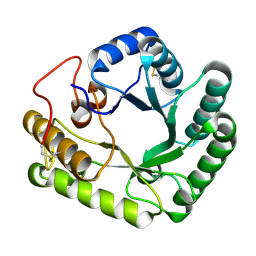 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6M62
 
 | |
7BR2
 
 | |
4HBN
 
 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
7JQB
 
 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
