4DGV
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, P2(1) form | | Descriptor: | E2 peptide, HCV1 Heavy Chain, HCV1 Light Chain, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DGY
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, C2 form | | Descriptor: | CHLORIDE ION, E2 peptide, GLYCEROL, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2012-01-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
4EW1
 
 | | High resolution structure of human glycinamide ribonucleotide transformylase in apo form. | | Descriptor: | PHOSPHATE ION, SULFATE ION, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
4EW3
 
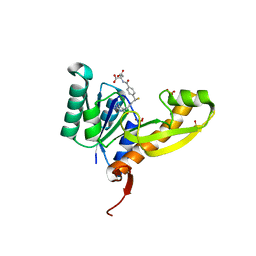 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10R-methylthio-DDATHF. | | Descriptor: | N-({4-[(1R)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
4EW2
 
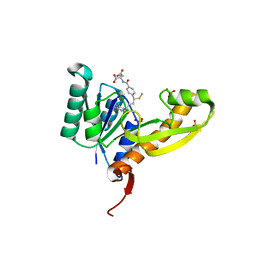 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10S-methylthio-DDATHF. | | Descriptor: | N-({4-[(1S)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
4EEF
 
 | |
6E3H
 
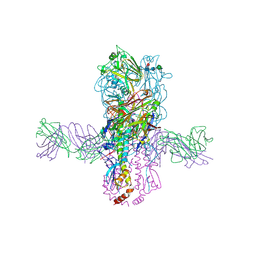 | | Crystal structure of S9-3-37 bound to H5 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2018-07-14 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The D3-9 gene segment encodes for recurring and adaptable binding motifs in broadly neutralizing antibodies to influenza virus
Cell Host Microbe, 2018
|
|
2R6Q
 
 | |
2R9U
 
 | |
6TZB
 
 | |
6UC5
 
 | | Fab397 in complex with NPNA peptide | | Descriptor: | Fab397 heavy chain, Fab397 light chain, NPNA peptide | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse Antibody Responses to Conserved Structural Motifs in Plasmodium falciparum Circumsporozoite Protein.
J.Mol.Biol., 432, 2020
|
|
7SBU
 
 | |
7T7B
 
 | |
7TP3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7U8M
 
 | |
7U8J
 
 | |
7U0K
 
 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7U8L
 
 | |
7TI6
 
 | |
7T1V
 
 | |
2IU3
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
