6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWT
 
 | |
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
8EQ5
 
 | | Crystal structure of the N-terminal kinase domain of RSK2 in complex with SPRED2 (131-160) | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3, Sprouty-related, ... | | Authors: | Bonsor, D.A, Lopez, J, McCormick, F, Simanshu, D.K. | | Deposit date: | 2022-10-07 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ribosomal S6 kinase 2 (RSK2)-SPRED2 complex regulates the phosphorylation of RSK substrates and MAPK signaling.
J.Biol.Chem., 299, 2023
|
|
6ZWZ
 
 | |
6ZX1
 
 | | OMPD-domain of human UMPS in complex with 6-Aza-UMP at 1.0 Angstroms resolution | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX0
 
 | | OMPD-domain of human UMPS in complex with the substrate OMP at 1.25 Angstroms resolution | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZWY
 
 | |
6ZX3
 
 | | OMPD-domain of human UMPS in complex with 6-thiocarboxamido-UMP at 1.15 Angstroms resolution | | Descriptor: | GLYCEROL, PROLINE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX2
 
 | | OMPD-domain of human UMPS in complex with 6-carboxamido-UMP at 1.2 Angstroms resolution | | Descriptor: | PROLINE, SULFATE ION, Uridine 5'-monophosphate synthase, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7AM9
 
 | | OMPD-domain of human UMPS in complex with the substrate OMP at 0.99 Angstroms resolution | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7ASQ
 
 | |
7B2N
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic Fructose bisphosphate aldolase | | Descriptor: | CHLORIDE ION, Fructose-bisphosphate aldolase 1, chloroplastic, ... | | Authors: | Le Moigne, T, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-11-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of chloroplast fructose-1,6-bisphosphate aldolase from the green alga Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2022
|
|
7CO1
 
 | | Crystal structure of SMAD2 in complex with wild-type CBP | | Descriptor: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
7OXZ
 
 | | VDR complex with a side-chain hydroxylated derivative of lithocholic acid | | Descriptor: | (3R,6R)-6-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]heptane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and evaluation of side-chain hydroxylated derivatives of lithocholic acid as potent agonists of the vitamin D receptor (VDR).
Bioorg.Chem., 115, 2021
|
|
7OY4
 
 | | VDR complex of a side-chain hydroxylated derivatives of lithocholic acid | | Descriptor: | (3S,6R)-6-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]heptane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and evaluation of side-chain hydroxylated derivatives of lithocholic acid as potent agonists of the vitamin D receptor (VDR).
Bioorg.Chem., 115, 2021
|
|
1F3J
 
 | | HISTOCOMPATIBILITY ANTIGEN I-AG7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, LYSOZYME C, ... | | Authors: | Latek, R.R, Unanue, E.R, Fremont, D.H. | | Deposit date: | 2000-06-04 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice.
Immunity, 12, 2000
|
|
1FD4
 
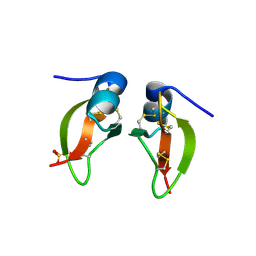 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
5DMI
 
 | |
1FD3
 
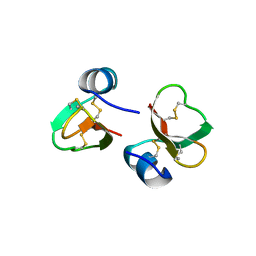 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
5DMJ
 
 | |
3LJE
 
 | | The X-ray structure of zebrafish RNase5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase5 | | Authors: | Russo Krauss, I, Merlino, A, Coscia, F, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3LN8
 
 | | The X-ray structure of Zf-RNase-1 from a new crystal form at pH 7.3 | | Descriptor: | HYDROLASE, SULFATE ION | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-02-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3LJD
 
 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
4YNK
 
 | | Crystal structure of vitamin D receptor ligand binding domain complexed with a 19-norvitamin D compound | | Descriptor: | (1R,3R,7E,17beta)-17-{(5S)-5-hydroxy-5-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-1-yl]penta-1,3-diyn-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Coactivator peptide drip from cDNA FLJ50196, highly similar to Peroxisome proliferator-activated receptor-binding protein, ... | | Authors: | Watarai, Y, Ikura, T, Ito, N. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, Biological Activities, and X-ray Crystal Structural Analysis of 25-Hydroxy-25(or 26)-adamantyl-17-[20(22),23-diynyl]-21-norvitamin D Compounds
J.Med.Chem., 58, 2015
|
|
