1EAE
 
 | |
1EAC
 
 | |
1EAB
 
 | |
1EAD
 
 | |
8DY2
 
 | | Crystal Structure of spFv GLK1 | | Descriptor: | SULFATE ION, spFv GLK1 LH | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY4
 
 | | Crystal Structure of spFv CAT2200 HL | | Descriptor: | spFv CAT2200 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY5
 
 | |
8DY0
 
 | | Crystal Structure of spFv GLK1 HL | | Descriptor: | spFv GLK1 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY3
 
 | | Crystal Structure of spFv GLK2 HL | | Descriptor: | SULFATE ION, spFv GLK2 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY1
 
 | |
7JGT
 
 | | Crystal Structure of FN3tt | | Descriptor: | Fibronectin type-III domain-containing protein | | Authors: | Luo, J, Malia, T.J. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
7JGU
 
 | | Structure of FN3tt mut | | Descriptor: | Fibronectin type-III domain-containing protein, PENTAETHYLENE GLYCOL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
2ASI
 
 | | ASPARTIC PROTEINASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTIC PROTEINASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Jia, Z, Vandonselaar, M, Kepliakov, P.S.A, Quail, J.W. | | Deposit date: | 1995-12-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 A resolution.
J.Mol.Biol., 268, 1997
|
|
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7PQN
 
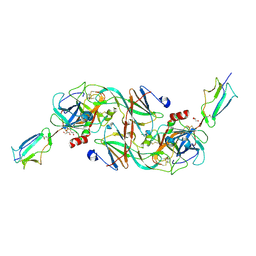 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
3I84
 
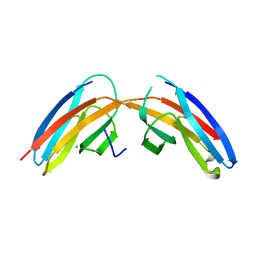 | |
3I85
 
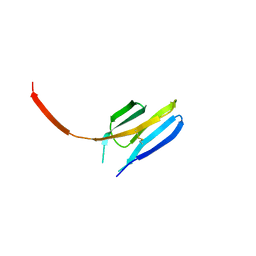 | |
3HMW
 
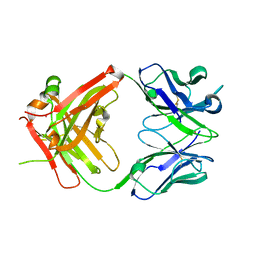 | | Crystal structure of ustekinumab FAB | | Descriptor: | CADMIUM ION, USTEKINUMAB FAB HEAVY CHAIN, USTEKINUMAB FAB LIGHT CHAIN | | Authors: | Luo, J. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
3HMX
 
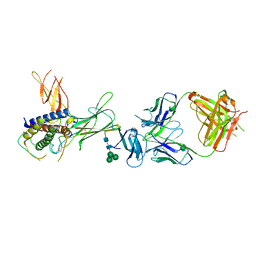 | | Crystal structure of ustekinumab FAB/IL-12 complex | | Descriptor: | Interleukin-12 subunit alpha, Interleukin-12 subunit beta, USTEKINUMAB FAB HEAVY CHAIN, ... | | Authors: | Luo, J. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
1KIF
 
 | | D-AMINO ACID OXIDASE FROM PIG KIDNEY | | Descriptor: | BENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Todone, F, Mattevi, A. | | Deposit date: | 1996-01-19 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5KF4
 
 | |
1N3B
 
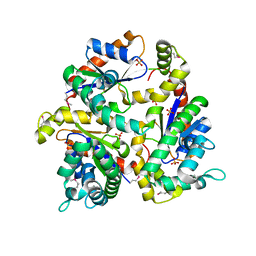 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
