4EZZ
 
 | |
4EZQ
 
 | |
4EZY
 
 | |
4EZN
 
 | |
4EZX
 
 | |
3QNJ
 
 | |
8B55
 
 | | Human ADGRG4 PTX-like domain | | Descriptor: | Adhesion G-protein coupled receptor G4, MAGNESIUM ION | | Authors: | Kieslich, B, Straeter, N. | | Deposit date: | 2022-09-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J.Biol.Chem., 299, 2023
|
|
8BFF
 
 | | Human PPARgamma in complex with MINCH bound to the AF-2 sub-pocket | | Descriptor: | (1~{S},2~{R})-2-[(4~{R})-4-methylheptoxy]carbonylcyclohexane-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Useini, A, Straeter, N. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the activation of PPAR gamma by the plasticizer metabolites MEHP and MINCH.
Environ Int, 173, 2023
|
|
8BF2
 
 | |
8BF1
 
 | |
8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | Descriptor: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | Authors: | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
4R3U
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
2BJ6
 
 | | Crystal Structure of a decameric HNA-RNA hybrid | | Descriptor: | 5'-R(*GP*GP*CP*AP*UP*UP*AP*CP*GP*GP)-3', SULFATE ION, SYNTHETIC HNA | | Authors: | Maier, T, Przylas, I, Straeter, N, Herdewijn, P, Saenger, W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reinforced Hna Backbone Hydration in the Crystal Structure of a Decameric Hna/RNA Hybrid
J.Am.Chem.Soc., 127, 2005
|
|
4HY9
 
 | |
4HYB
 
 | |
6SJU
 
 | |
6SHI
 
 | |
6SHH
 
 | |
4E81
 
 | |
4JWI
 
 | |
4JWC
 
 | |
4H2B
 
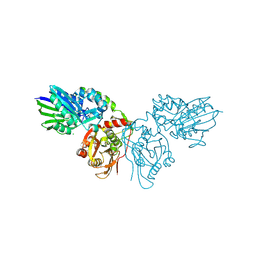 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with Baicalin | | Descriptor: | 5'-nucleotidase, 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, CALCIUM ION, ... | | Authors: | Zebisch, M, Pippel, J, Knapp, K, Straeter, N. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
3ZX2
 
 | | NTPDase1 in complex with Decavanadate | | Descriptor: | ACETIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
4H1Y
 
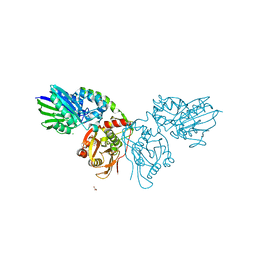 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with PSB11552 | | Descriptor: | 2,2'-[(2-{[2-({[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)ethyl]amino}-2-oxoethyl)imino]diacetic acid (non-preferred name), 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Pippel, J, Zebisch, M, Knapp, K, Straeter, N. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
4JWE
 
 | |
