8X4H
 
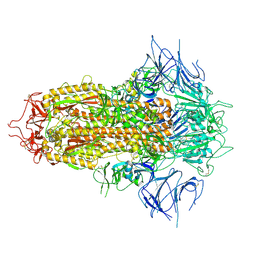 | | SARS-CoV-2 JN.1 Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5Q
 
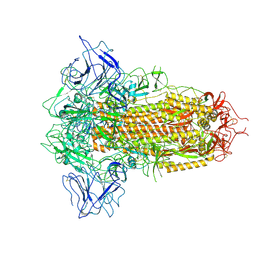 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHV
 
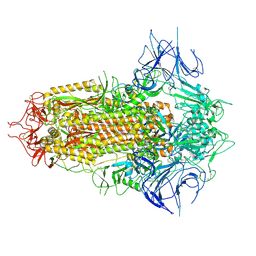 | | Spike Trimer of BA.2.86 with three RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHS
 
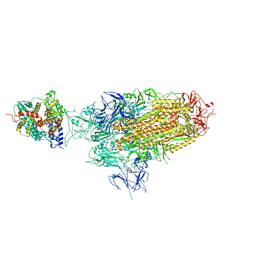 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUS
 
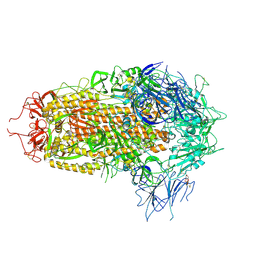 | | JN.1 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUR
 
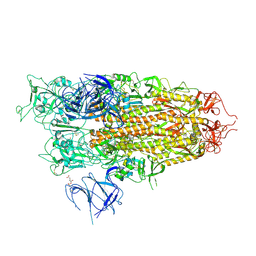 | | BA.2.86 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUU
 
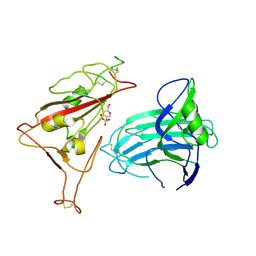 | | BA.2.86-T356K Spike Trimer in complex with heparan sulfate (Local refinement) | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUT
 
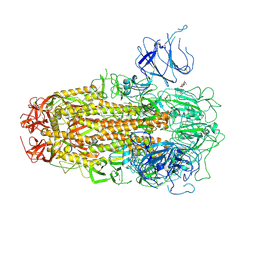 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P, Mao, X. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
7VS3
 
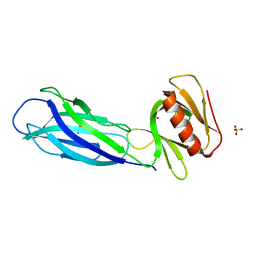 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | Descriptor: | Calcium-dependent secretion activator 1, SULFATE ION | | Authors: | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
6V3W
 
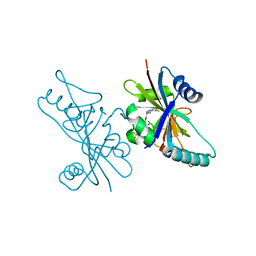 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment with four point mutations in complex with RBN-2397 | | Descriptor: | 5-{[(2S)-1-(3-oxo-3-{4-[5-(trifluoromethyl)pyrimidin-2-yl]piperazin-1-yl}propoxy)propan-2-yl]amino}-4-(trifluoromethyl)pyridazin-3(2H)-one, CHLORIDE ION, Protein mono-ADP-ribosyltransferase PARP12 | | Authors: | Swinger, K.K, Gozgit, J.M, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity.
Cancer Cell, 39, 2021
|
|
6W65
 
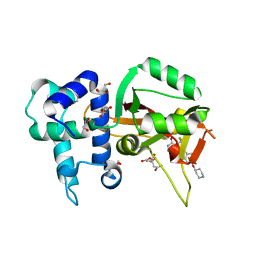 | | Human PARP16 in complex with RBN010860 | | Descriptor: | 1,2-ETHANEDIOL, 5-{5-[(piperidin-4-yl)oxy]-2H-isoindol-2-yl}-4-(trifluoromethyl)pyridazin-3(2H)-one, CITRIC ACID, ... | | Authors: | Swinger, K.K, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | In Vitro and Cellular Probes to Study PARP Enzyme Target Engagement.
Cell Chem Biol, 27, 2020
|
|
4TZU
 
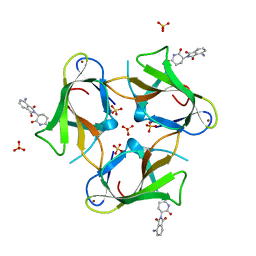 | | Crystal Structure of Murine Cereblon in Complex with Pomalidomide | | Descriptor: | Protein cereblon, S-Pomalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
7LUN
 
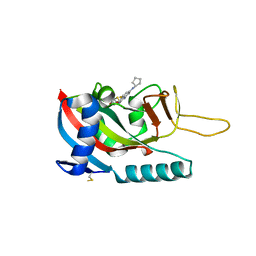 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN011980 | | Descriptor: | 7-(cyclopentylamino)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
7L9Y
 
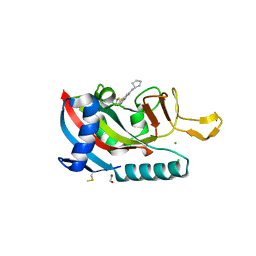 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012042 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopentylamino)-5-fluoro-2-{[(piperidin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
7Y43
 
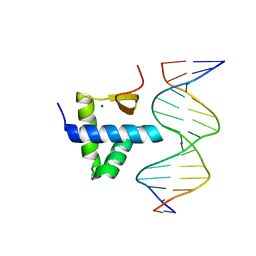 | | Crystal structure of the KAT6A WH domain and its bound double stranded DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*TP*GP*CP*GP*CP*AP*CP*TP*CP*C)-3'), Histone acetyltransferase KAT6A, MAGNESIUM ION | | Authors: | Wang, Z, Jia, Y. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The histone acetyltransferase KAT6A is recruited to unmethylated CpG islands via a DNA binding winged helix domain.
Nucleic Acids Res., 51, 2023
|
|
7CNU
 
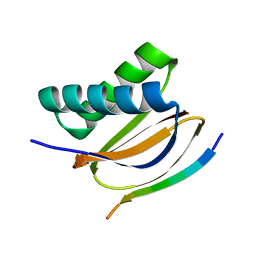 | | Crystal structure of DLC2 in complex with BMF peptide | | Descriptor: | Bcl-2-modifying factor, Dynein light chain 2, cytoplasmic | | Authors: | Wen, Y, Shao, Y. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-canonical phosphorylation of Bmf by p38 MAPK promotes its apoptotic activity in anoikis.
Cell Death Differ., 29, 2022
|
|
7EGF
 
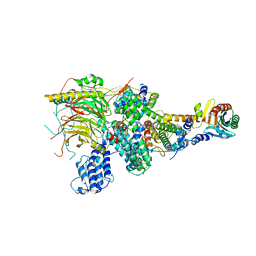 | | TFIID lobe A subcomplex | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 11, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 2021
|
|
7EGA
 
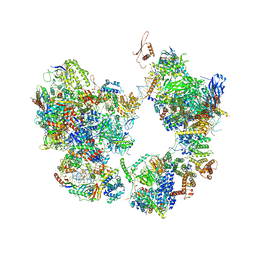 | | TFIID-based intermediate PIC on PUMA promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EDX
 
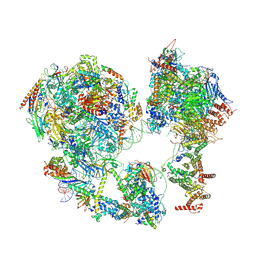 | | p53-bound TFIID-based core PIC on HDM2 promoter | | Descriptor: | DNA (84-mer), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGG
 
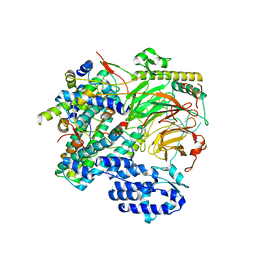 | | TFIID lobe B subcomplex | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 4, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGB
 
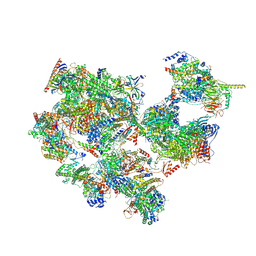 | | TFIID-based holo PIC on SCP promoter | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG7
 
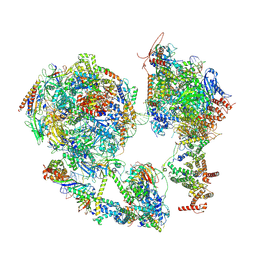 | | TFIID-based core PIC on SCP promoter | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGH
 
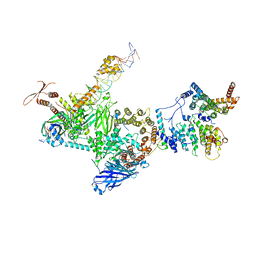 | | TFIID lobe C subcomplex | | Descriptor: | DNA (45-MER), Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGI
 
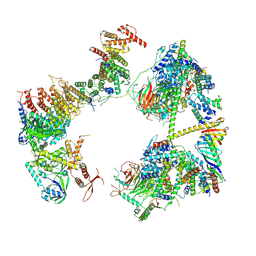 | | TFIID in rearranged conformation | | Descriptor: | TATA-box-binding protein, Transcription initiation factor IIA subunit 1, Transcription initiation factor IIA subunit 2, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (9.82 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG9
 
 | | TFIID-based intermediate PIC on SCP promoter | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
