7Y5Z
 
 | | CryoEM structure of human PS2-containing gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5T
 
 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
8HFI
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant desipramine in an inward-open state at resolution of 2.5 angstrom. | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFF
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of norepinephrine in an inward-open state at resolution of 2.9 angstrom. | | Descriptor: | L-NOREPINEPHRINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFL
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant bupropion in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | Bupropion, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFG
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of dopamine in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | CHLORIDE ION, L-DOPAMINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFE
 
 | | Cryo-EM structure of human norepinephrine transporter NET in an inward-open state at resolution of 2.5 angstrom | | Descriptor: | CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
3JA8
 
 | | Cryo-EM structure of the MCM2-7 double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | Authors: | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
8IZT
 
 | |
8IZU
 
 | |
7YBG
 
 | |
8HDA
 
 | |
8ZAI
 
 | |
8Z1L
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8KCP
 
 | | Cryo-EM structure of human gamma-secretase in complex with Crenigacestat | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KCO
 
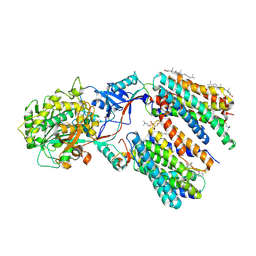 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KCU
 
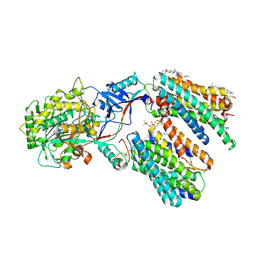 | | Cryo-EM structure of human gamma-secretase in complex with MK-0752 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KCT
 
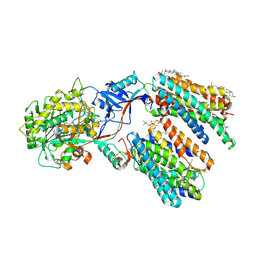 | | Cryo-EM structure of human gamma-secretase in complex with Nirogacestat | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KCS
 
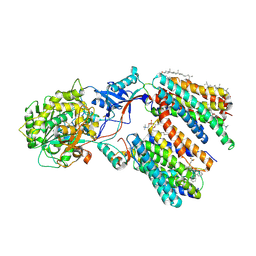 | | Cryo-EM structure of human gamma-secretase in complex with BMS906024 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4CSU
 
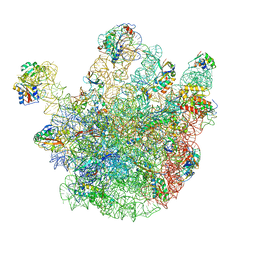 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
5YLZ
 
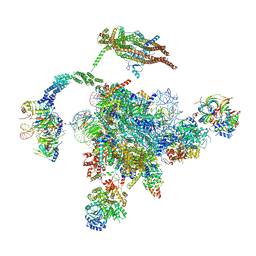 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
5Y88
 
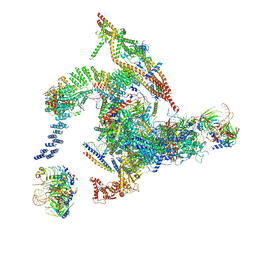 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
6A70
 
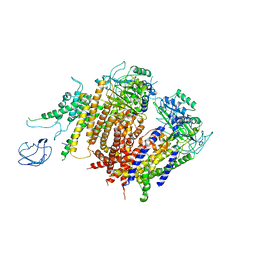 | | Structure of the human PKD1/PKD2 complex | | Descriptor: | Polycystin-1, Polycystin-2 | | Authors: | Su, Q, Hu, F, Ge, X, Lei, J, Yu, S, Wang, T, Zhou, Q, Mei, C, Shi, Y. | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human PKD1-PKD2 complex.
Science, 361, 2018
|
|
5Z57
 
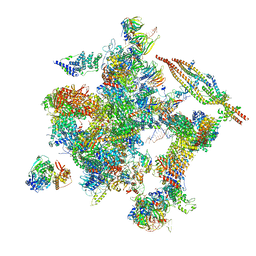 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
