7CHN
 
 | | Crystal structure of TTK kinase domain in complex with compound 9 | | Descriptor: | 4-(cyclohexylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CLH
 
 | | Crystal structure of TTK kinase domain in complex with compound 19 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(methylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CJA
 
 | | Crystal structure of TTK kinase domain in complex with compound 28 | | Descriptor: | 4-(cyclopentylmethylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-09 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CIL
 
 | | Crystal structure of TTK kinase domain in complex with compound 7 | | Descriptor: | 4-(cyclohexylamino)-2-[(1-methylpyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
6IBL
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
7AD3
 
 | | Class D GPCR Ste2 dimer coupled to two G proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-factor mating pheromone, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Velazhahan, V, Tate, C. | | Deposit date: | 2020-09-14 | | Release date: | 2020-12-09 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the class D GPCR Ste2 dimer coupled to two G proteins.
Nature, 589, 2021
|
|
4LUY
 
 | | Crystal structure of CdALR mutant K 271 T | | Descriptor: | Alanine racemase | | Authors: | Asojo, O.A. | | Deposit date: | 2013-07-25 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses of alanine racemase from the multidrug-resistant Clostridium difficile strain 630.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7XM8
 
 | | Glucagon amyloid fibril | | Descriptor: | Glucagon | | Authors: | Jeong, H, Lin, Y, Lee, Y.-H. | | Deposit date: | 2022-04-25 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomistic zipper-like amyloid structure of full-length glucagon
To Be Published
|
|
6X6N
 
 | |
3NJ4
 
 | | Fluoro-neplanocin A in Human S-Adenosylhomocysteine Hydrolase | | Descriptor: | (4S,5S)-4-(6-amino-9H-purin-9-yl)-3-fluoro-5-hydroxy-2-(hydroxymethyl)cyclopent-2-en-1-one, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jeong, L.S, Lee, K.M, Hwang, K.Y, Choi, S, Heo, Y.S. | | Deposit date: | 2010-06-17 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure and binding mode analysis of human S-adenosylhomocysteine hydrolase complexed with novel mechanism-based inhibitors, haloneplanocin A analogues.
J.Med.Chem., 54, 2011
|
|
2RSY
 
 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
8IVU
 
 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
3EGM
 
 | | Structural basis of iron transport gating in Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Shin, H.J, Lee, J.H. | | Deposit date: | 2008-09-11 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of ferritin from Helicobacter pylori reveals unusual conformational changes for iron uptake.
J.Mol.Biol., 390, 2009
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
6A6G
 
 | | Crystal structure of thermostable FiSufS-SufU complex from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | Cysteine desulfurase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
6A6F
 
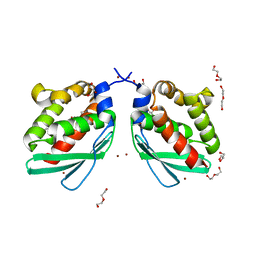 | | Crystal structure of Putative iron-sulfur cluster assembly scaffold protein for SUF system (FiSufU) from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Iron-sulfur cluster assembly scaffold protein NifU, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
6A6E
 
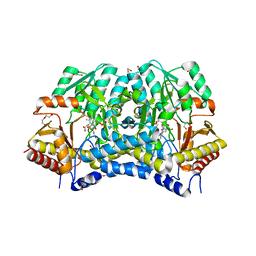 | | Crystal structure of thermostable Cysteine desulfurase (FiSufS) from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | CITRIC ACID, Cysteine desulfurase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
5GWO
 
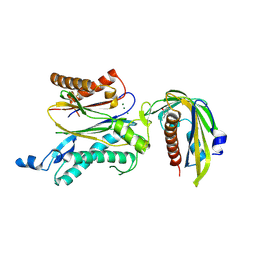 | | Crystal structure of RCAR3:PP2C S265F/I267M with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
5GWP
 
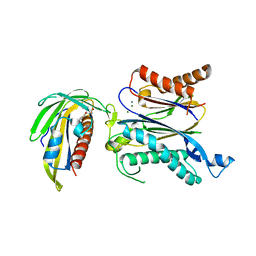 | | Crystal structure of RCAR3:PP2C wild-type with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
7WCG
 
 | |
6JPV
 
 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15000653 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
6K39
 
 | | Structural analysis of AIMP2-DX2 and HSP70 interaction | | Descriptor: | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 | | Authors: | Cho, H.Y, Son, S.Y, Jeon, Y.H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3981427 Å) | | Cite: | Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
Nat.Chem.Biol., 16, 2020
|
|
7E64
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | Descriptor: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | Descriptor: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
