3LED
 
 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009 | | Descriptor: | 3-oxoacyl-acyl carrier protein synthase III, FORMIC ACID | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-14 | | Release date: | 2010-01-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009
To be Published
|
|
3LLB
 
 | | The crystal structure of the protein PA3983 with unknown function from Pseudomonas aeruginosa PAO1 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Kagan, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the protein NE1376 with unknown function from Nitrosomonas europaea ATCC 19718
To be Published
|
|
3LHH
 
 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
3LEQ
 
 | | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A | | Descriptor: | uncharacterized protein cvnB5 | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A
To be Published
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8DIL
 
 | | Crystal structure of putative nitroreductase from Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Skarina, T, Mesa, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative nitroreductase from Salmonella enterica
to be published
|
|
8EBG
 
 | | Crystal structure of the probable FhuD FeIII-dicitrate-binding domain protein FecB from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, FEIII-dicitrate-binding periplasmic lipoprotein FecB, FORMIC ACID, ... | | Authors: | Cuff, M, Kim, Y, Endres, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of the probable FhuD FeIII-dicitrate-binding domain protein FecB from Mycobacterium tuberculosis
To Be Published
|
|
8GHX
 
 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | Descriptor: | 1,2-ETHANEDIOL, Cellulase CelD | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8GHY
 
 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | Descriptor: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8EBC
 
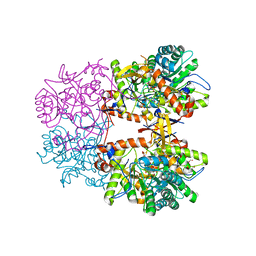 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | Descriptor: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
8CRV
 
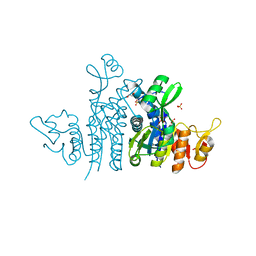 | | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Carbamate kinase, FORMIC ACID, ... | | Authors: | Kim, Y, Skarina, T, Mesa, N, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa
To Be Published
|
|
5KIN
 
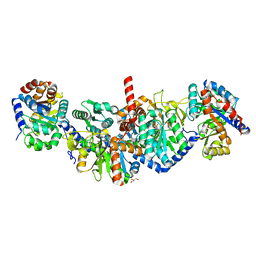 | | Crystal structure of tryptophan synthase alpha beta complex from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, Tryptophan synthase alpha chain, Tryptophan synthase beta chain | | Authors: | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-16 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conservation of the structure and function of bacterial tryptophan synthases.
Iucrj, 6, 2019
|
|
8TTP
 
 | | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-14 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam
to be published
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
8U00
 
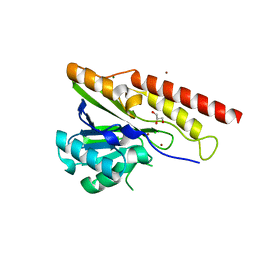 | | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides
To Be Published
|
|
8U12
 
 | |
3LK7
 
 | | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylmuramoylalanine--D-glutamate ligase | | Authors: | Stein, A.J, Sather, A, Shakelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A
To be Published
|
|
3LLV
 
 | | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A | | Descriptor: | Exopolyphosphatase-related protein, PHOSPHATE ION | | Authors: | Stein, A.J, Chang, C, Weger, A, Hendricks, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A
To be Published
|
|
3LOQ
 
 | | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tan, K, Weger, A, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-16 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | Authors: | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-05-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
4XXT
 
 | | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fusion of predicted Zn-dependent amidase/peptidase (Cell wall hydrolase/DD-carboxypeptidase family) and uncharacterized domain of ErfK family peptodoglycan-binding domain, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from from Clostridium acetobutylicum ATCC 824
To Be Published
|
|
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
5IX8
 
 | | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822 | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transport system, substrate-binding protein, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-06 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822
To Be Published
|
|
3BV8
 
 | | Crystal structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SODIUM ION, Tetrahydrodipicolinate acetyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus.
TO BE PUBLISHED
|
|
