2G0U
 
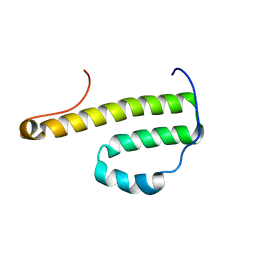 | | Solution Structure of Monomeric BsaL, the Type III Secretion Needle Protein of Burkholderia pseudomallei | | Descriptor: | type III secretion system needle protein | | Authors: | Zhang, L, Wang, Y, Picking, W.L, Picking, W.D, De Guzman, R.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Monomeric BsaL, the Type III Secretion Needle Protein of Burkholderia pseudomallei.
J.Mol.Biol., 359, 2006
|
|
8JU8
 
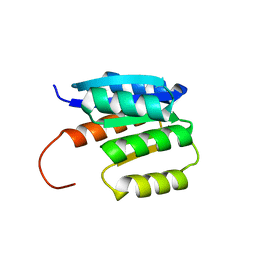 | | de novo designed protein | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2023-06-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | de novo designed Rossmann fold protein
To Be Published
|
|
2H1Y
 
 | |
6AUE
 
 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-b | | Descriptor: | N-biotin-C-Co4(mu3-O)4(OAc)(Py)4(H2O)3-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
4I5X
 
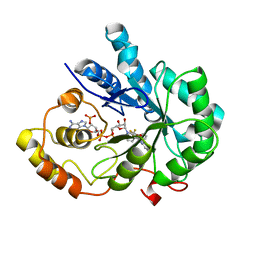 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
6AUL
 
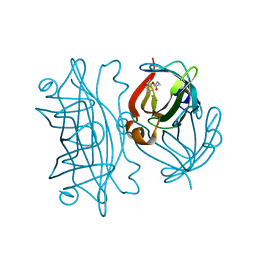 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-b | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
4GQG
 
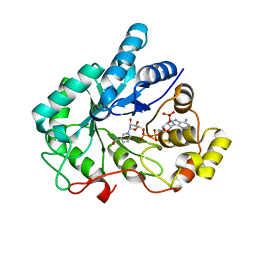 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
6AUC
 
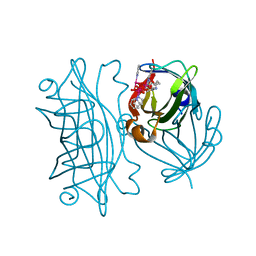 | | Artificial metalloproteins containing a Co4O4 active site - 2xm-Sav | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)4(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurackal, J, Huerta-Lavorie, R, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
3UOB
 
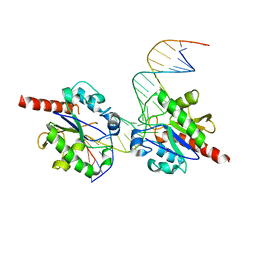 | |
6AUH
 
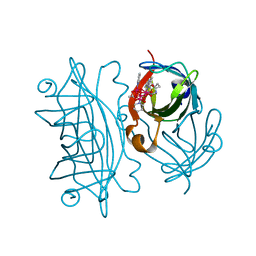 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-a | | Descriptor: | N-biotin-C-Co4(mu3-O)4(OAc)(Py)3(H2O)3-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
3UO7
 
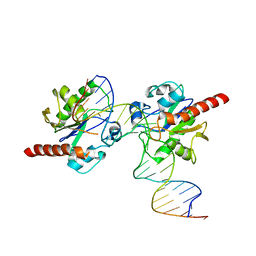 | | Crystal structure of Human Thymine DNA Glycosylase Bound to Substrate 5-carboxylcytosine | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*GP*TP*AP*CP*AP*TP*GP*AP*GP*CP*AP*GP*TP*GP*GP*A)-3', 5'-D(*CP*CP*AP*CP*TP*GP*CP*TP*CP*AP*(1CC)P*GP*TP*AP*CP*AP*GP*AP*GP*CP*TP*GP*T)-3', G/T mismatch-specific thymine DNA glycosylase | | Authors: | Zhang, L, He, C. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA.
Nat.Chem.Biol., 8, 2012
|
|
6AUO
 
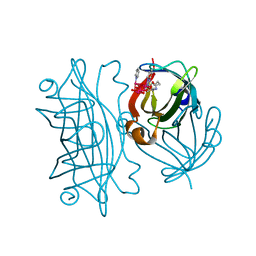 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112F | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)3(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
1E31
 
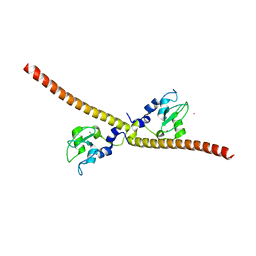 | | SURVIVIN DIMER H. SAPIENS | | Descriptor: | APOPTOSIS INHIBITOR SURVIVIN, COBALT (II) ION, ZINC ION | | Authors: | Chantalat, L, Skoufias, D.A, Margolis, R.L, Dideberg, O. | | Deposit date: | 2000-06-04 | | Release date: | 2001-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of Human Survivin Reveals a Bow Tie-Shaped Dimer with Two Unusual Alpha-Helical Extensions
Mol.Cell, 6, 2000
|
|
1DXK
 
 | |
4JIH
 
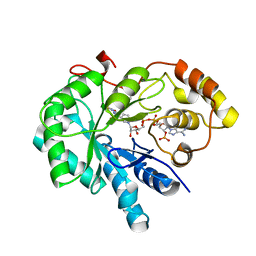 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Epalrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-[(2E)-2-methyl-3-phenylprop-2-en-1-ylidene]-4-oxo-2-thioxo-1,3-thiazolidin-3-yl}acetic acid | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JIR
 
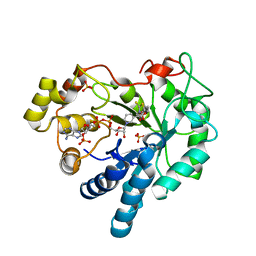 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Epalrestat | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X. | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JII
 
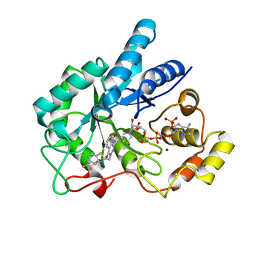 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
7JZY
 
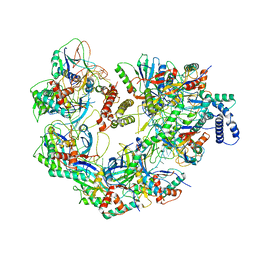 | | CryoEM structure of a CRISPR-Cas complex | | Descriptor: | AcrF9, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of a CRISPR-Cas complex
To Be Published
|
|
1U9U
 
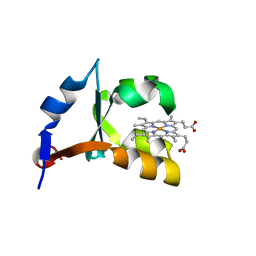 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2P87
 
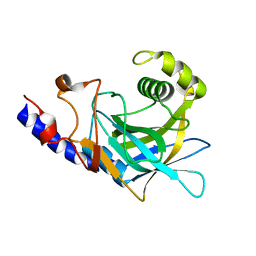 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
6IPB
 
 | |
4IF2
 
 | |
1U9M
 
 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1KXK
 
 | |
4J0W
 
 | | Structure of U3-55K | | Descriptor: | U3 small nucleolar RNA-interacting protein 2 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
