1V82
 
 | | Crystal structure of human GlcAT-P apo form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V83
 
 | | Crystal structure of human GlcAT-P in complex with Udp and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V84
 
 | | Crystal structure of human GlcAT-P in complex with N-acetyllactosamine, Udp, and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
2ZBG
 
 | | Calcium pump crystal structure with bound AlF4 and TG in the absence of calcium | | Descriptor: | MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ZBF
 
 | | Calcium pump crystal structure with bound BeF3 and TG in the absence of calcium | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ZBE
 
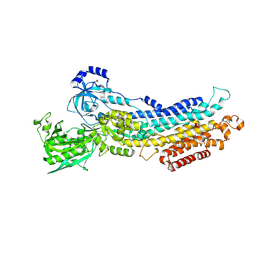 | |
2ZFA
 
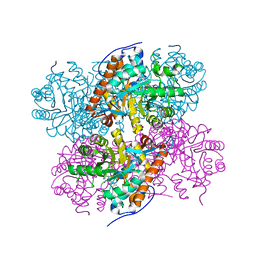 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
3A5T
 
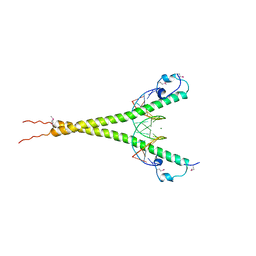 | | Crystal structure of MafG-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
2ZBD
 
 | | Crystal Structure of the SR Calcium Pump with Bound Aluminium Fluoride, ADP and Calcium | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
2ZXE
 
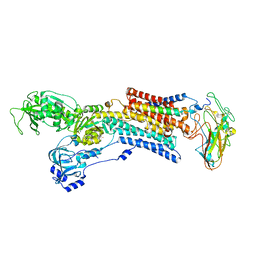 | | Crystal structure of the sodium - potassium pump in the E2.2K+.Pi state | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Shinoda, T, Ogawa, H, Cornelius, F, Toyoshima, C. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the sodium - potassium pump at 2.4 A resolution
Nature, 459, 2009
|
|
2ZE5
 
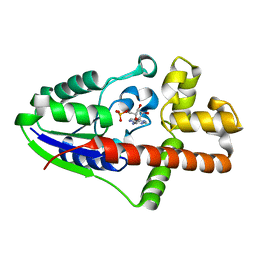 | | Crystal Structure of adenosine phosphate-isopentenyltransferase | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Isopentenyl transferase | | Authors: | Sakakibara, H. | | Deposit date: | 2007-12-06 | | Release date: | 2008-02-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural insight into the reaction mechanism and evolution of cytokinin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZE6
 
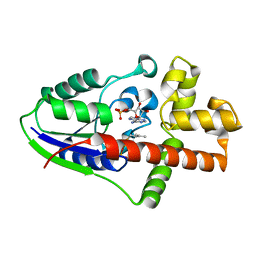 | |
2ZE8
 
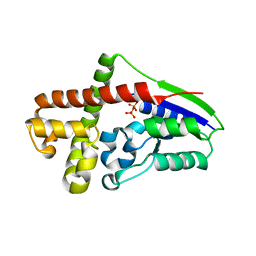 | |
2ZE7
 
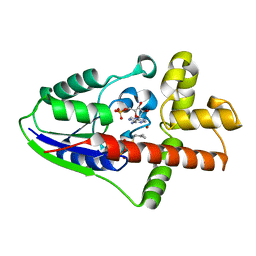 | |
3NNC
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NNA
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NNH
 
 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NMR
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-22 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
1D2G
 
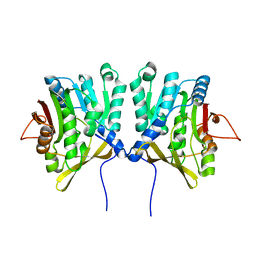 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE FROM RAT LIVER | | Descriptor: | GLYCINE N-METHYLTRANSFERASE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-08 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2C
 
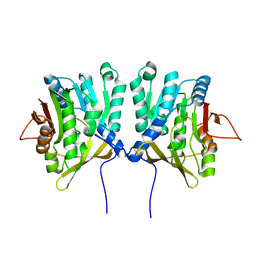 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2H
 
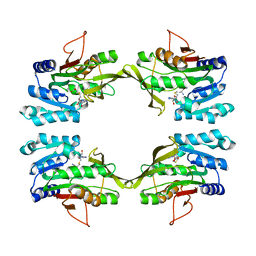 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLHOMOCYSTEINE | | Descriptor: | GLYCINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-11 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
4G78
 
 | |
3VX8
 
 | |
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
6OVA
 
 | | Crystal Structure of TYK2 with novel pyrrolidinone inhibitor | | Descriptor: | 6-({4-[(3S)-3-cyano-3-cyclopropyl-2-oxopyrrolidin-1-yl]pyridin-2-yl}amino)-N,N-dimethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Skene, R.J, Hoffman, I.D. | | Deposit date: | 2019-05-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Efficient synthesis of tert-butyl 3-cyano-3-cyclopropyl-2-oxopyrrolidine-4-carboxylates: Highly functionalized 2-pyrrolidinone enabling access to novel macrocyclic Tyk2 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
