1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2N
 
 | |
1N6A
 
 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6C
 
 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N4M
 
 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
8ZPH
 
 | | SFX reaction state structure (40-60min) of alanine racemase | | Descriptor: | ALANINE, Alanine racemase 2, CHLORIDE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2024-05-30 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the reaction dynamics of alanine racemase using serial femtosecond crystallography.
Sci Rep, 14, 2024
|
|
8ZPG
 
 | | SFX reaction state structure (20-40min) of alanine racemase | | Descriptor: | ALANINE, Alanine racemase 2, CHLORIDE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2024-05-30 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the reaction dynamics of alanine racemase using serial femtosecond crystallography.
Sci Rep, 14, 2024
|
|
8ZPF
 
 | | SFX reaction state structure (0-20min) of alanine racemase | | Descriptor: | ALANINE, Alanine racemase 2, CHLORIDE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2024-05-30 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the reaction dynamics of alanine racemase using serial femtosecond crystallography.
Sci Rep, 14, 2024
|
|
8ZPE
 
 | |
8HST
 
 | |
8HSV
 
 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1YUC
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to Phospholipid and a Fragment of Human SHP | | Descriptor: | GLYCEROL, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor 0B2, ... | | Authors: | Ortlund, E.A, Yoonkwang, L, Solomon, I.H, Hager, J.M, Safi, R, Choi, Y, Guan, Z, Tripathy, A, Raetz, C.R.H, McDonnell, D.P, Moore, D.D, Redinbo, M.R. | | Deposit date: | 2005-02-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP
Nat.Struct.Mol.Biol., 12, 2005
|
|
2WZQ
 
 | | Insertion Mutant E173GP174 of the NS3 protease-helicase from dengue virus | | Descriptor: | CHLORIDE ION, GLYCEROL, NS3 PROTEASE-HELICASE | | Authors: | Luo, D, Wei, N, Doan, D, Paradkar, P, Chong, Y, Davidson, A, Kotaka, M, Lescar, J, Vasudevan, S. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility between the Protease and Helicase Domains of the Dengue Virus Ns3 Protein Conferred by the Linker Region and its Functional Implications.
J.Biol.Chem., 285, 2010
|
|
2B1I
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
1LB5
 
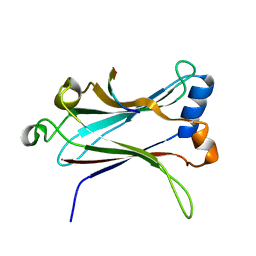 | | TRAF6-RANK Complex | | Descriptor: | TNF receptor-associated factor 6, receptor activator of nuclear factor-kappa B | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB4
 
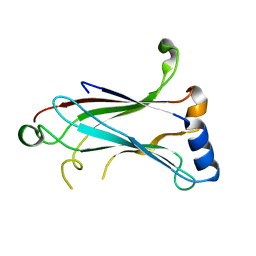 | | TRAF6 apo structure | | Descriptor: | TNF receptor-associated factor 6 | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB6
 
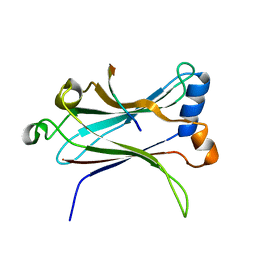 | | TRAF6-CD40 Complex | | Descriptor: | CD40 antigen, TNF receptor-associated factor 6 | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1Z8G
 
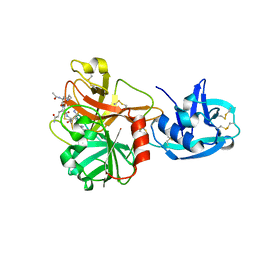 | | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. | | Descriptor: | ACE-LYS-GLN-LEU-ARG-Chloromethylketone, Serine protease hepsin | | Authors: | Herter, S, Piper, D.E, Aaron, W, Gabriele, T, Cutler, G, Cao, P, Bhatt, A.S, Choe, Y, Craik, C.S, Walker, N, Meininger, D, Hoey, T, Austin, R.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers
Biochem.J., 390, 2005
|
|
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
1Q90
 
 | | Structure of the cytochrome b6f (plastohydroquinone : plastocyanin oxidoreductase) from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Stroebel, D, Choquet, Y, Popot, J.-L, Picot, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Atypical Haem in the Cytochrome B6F Complex
Nature, 426, 2003
|
|
2IU3
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
