7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6G3D
 
 | | Crystal structure of Native EDDS lyase | | Descriptor: | Argininosuccinate lyase | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3E
 
 | | Crystal structure of EDDS lyase in complex with formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
1PZV
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
7MCE
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol | | Descriptor: | 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
6G3I
 
 | | Crystal structure of EDDS lyase in complex with N-(2-aminoethyl)aspartic acid (AEAA) | | Descriptor: | (2~{S})-2-(2-azanylethylamino)butanedioic acid, Argininosuccinate lyase, FUMARIC ACID | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3F
 
 | | Crystal structure of EDDS lyase in complex with fumarate | | Descriptor: | Argininosuccinate lyase, DI(HYDROXYETHYL)ETHER, FUMARIC ACID | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3H
 
 | | Crystal structure of EDDS lyase in complex with SS-EDDS | | Descriptor: | (2~{S})-2-[2-[[(2~{S})-1,4-bis(oxidanyl)-1,4-bis(oxidanylidene)butan-2-yl]amino]ethylamino]butanedioic acid, Argininosuccinate lyase | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3G
 
 | | Crystal structure of EDDS lyase in complex with succinate | | Descriptor: | Argininosuccinate lyase, DI(HYDROXYETHYL)ETHER, SUCCINIC ACID | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
7W26
 
 | | monolignol ferulate transferase | | Descriptor: | Ferulate monolignol transferase | | Authors: | Xi, L, Shuliu, D, Yue, F, Yi, Z. | | Deposit date: | 2021-11-22 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of the plant feruloyl-coenzyme A monolignol transferase provides insights into the formation of monolignol ferulate conjugates.
Biochem.Biophys.Res.Commun., 594, 2022
|
|
6PIB
 
 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PH9
 
 | | Crystal Structure of the Klebsiella pneumoniae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PJ3
 
 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SJ7
 
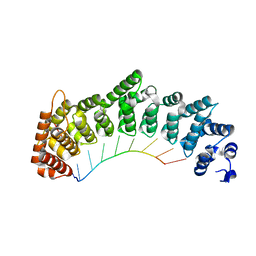 | |
7JUO
 
 | | CBP bromodomain complexed with YF2-23 | | Descriptor: | CREB-binding protein, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Principe, D, Li, Y, Huang, F, Rana, A, Thatcher, G. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | XP-524 is a dual-BET/EP300 inhibitor that represses oncogenic KRAS and potentiates immune checkpoint inhibition in pancreatic cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VAH
 
 | |
3B5H
 
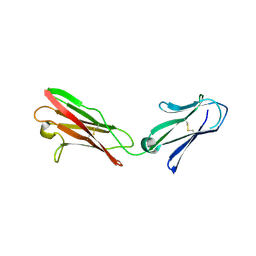 | |
4DNW
 
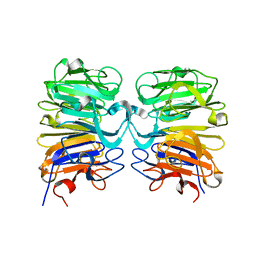 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
1GOL
 
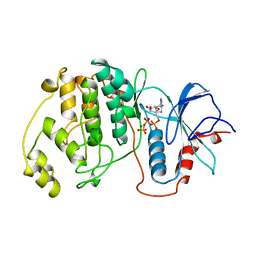 | |
6LPF
 
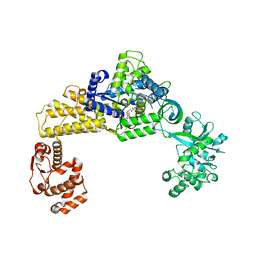 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
7D4B
 
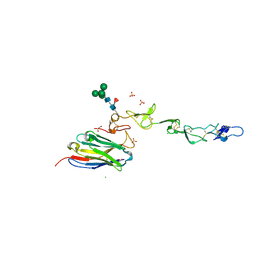 | | Crystal structure of 4-1BB in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
8K1C
 
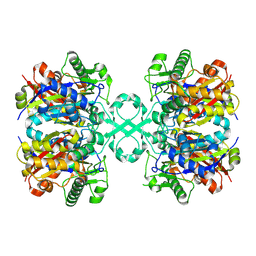 | |
8K1F
 
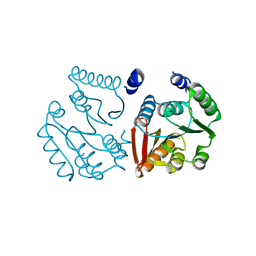 | |
6V6M
 
 | | Crystal structure of an inactive state of GMPPNP-bound RhoA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lin, Y, Zheng, Y. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of an inactive conformation of GTP-bound RhoA GTPase.
Structure, 29, 2021
|
|
7CZD
 
 | | Crystal structure of PD-L1 in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Programmed cell death 1 ligand 1, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
