4O8O
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to alpha-L-arabinose | | Descriptor: | Alpha-L-arabinofuranosidase, CALCIUM ION, alpha-L-arabinofuranose | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
4H4S
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
5LRJ
 
 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin C complex | | Descriptor: | Anabaenopeptin C, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
5LLO
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 3-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hy-droxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
1XZQ
 
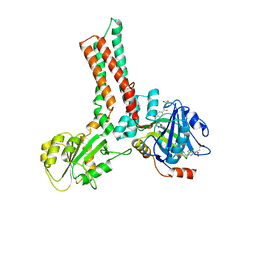 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima complexed with 5-formyl-THF | | Descriptor: | N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, Probable tRNA modification GTPase trmE | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
7WST
 
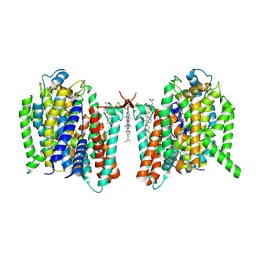 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-DMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]azetidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7WSU
 
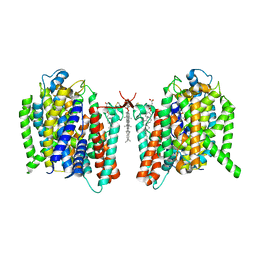 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-PDMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]pyrrolidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
6O9E
 
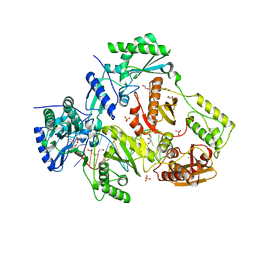 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA and INDOPY-1 | | Descriptor: | 5-methyl-1-(4-nitrophenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruiz, F.X, Hoang, A, Das, K, Arnold, E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1.
J.Med.Chem., 62, 2019
|
|
5LS9
 
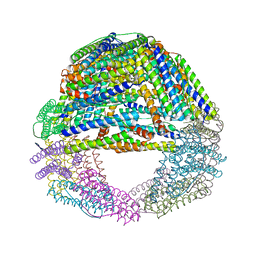 | | Humanized Archaeal ferritin | | Descriptor: | Ferritin, putative, MAGNESIUM ION | | Authors: | Baiocco, P, Trabuco, M.C, Boffi, A. | | Deposit date: | 2016-08-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Humanized archaeal ferritin as a tool for cell targeted delivery.
Nanoscale, 9, 2017
|
|
6GMP
 
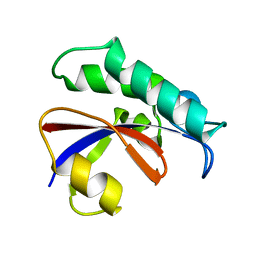 | | CRYSTAL STRUCTURE OF THE PPIASE DOMAIN OF TBPAR42 | | Descriptor: | PARVULIN 42 | | Authors: | Hoenig, D, Rute, A, Hofmann, E, Bayer, P, Gasper, R. | | Deposit date: | 2018-05-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of the 42 kDa Parvulin ofTrypanosoma brucei.
Biomolecules, 9, 2019
|
|
7WRR
 
 | |
7WSR
 
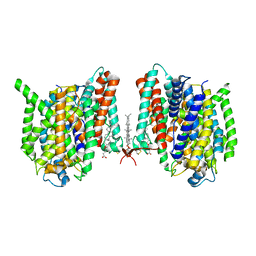 | | Cryo-EM structure of the barley Yellow stripe 1 transporter | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Iron-phytosiderophore transporter | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
1XXR
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus Nigra in complex with mannose | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
1ZRZ
 
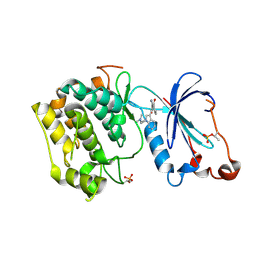 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
6OFQ
 
 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori in complex with peptide STSA | | Descriptor: | Heme-binding protein A / AI-2 binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-03-31 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
6GXS
 
 | | Crystal structure of CV39L lectin from Chromobacterium violaceum at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CV39L lectin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sykorova, P, Novotna, J, Demo, G, Pompidor, G, Dubska, E, Komarek, J, Fujdiarova, E, Haronikova, L, Varrot, A, Imberty, A, Shilova, N, Bovin, N, Pokorna, M, Wimmerova, M. | | Deposit date: | 2018-06-27 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of novel lectins from Burkholderia pseudomallei and Chromobacterium violaceum with seven-bladed beta-propeller fold.
Int.J.Biol.Macromol., 152, 2020
|
|
7W9Q
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
4LPU
 
 | |
6GYF
 
 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, ... | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
7W3E
 
 | | Bovine cytochrome c oxidese in CN-bound fully reduced state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
4H4U
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 T176R mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
7W9G
 
 | | Complex structure of Mpro with ebselen-derivative inhibitor | | Descriptor: | 3C-like proteinase nsp5, SELENIUM ATOM | | Authors: | Sahoo, P, Kumar, A. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7W9R
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-18 | | Descriptor: | (2~{R})-2-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
6OTA
 
 | |
4H6U
 
 | | Tubulin acetyltransferase mutant | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, PHOSPHATE ION, ... | | Authors: | Roll-Mecak, A, Kizub, V, Szyk, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4482 Å) | | Cite: | Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus.
J.Biol.Chem., 287, 2012
|
|
