6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
6XKL
 
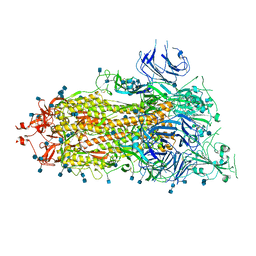 | | SARS-CoV-2 HexaPro S One RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrapp, D, Hsieh, C.-L, Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.
Science, 369, 2020
|
|
7U0A
 
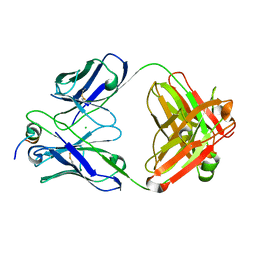 | |
7U09
 
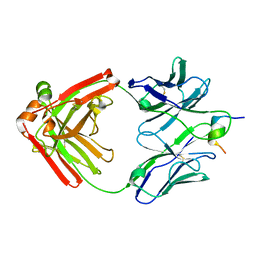 | |
7U0E
 
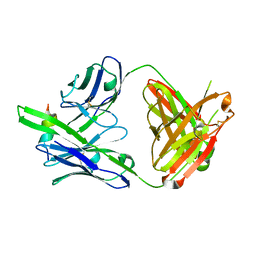 | |
6NM4
 
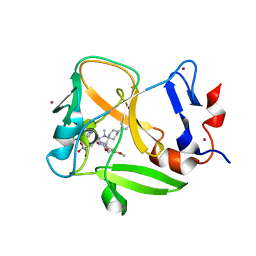 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | Descriptor: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | Authors: | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
7SL5
 
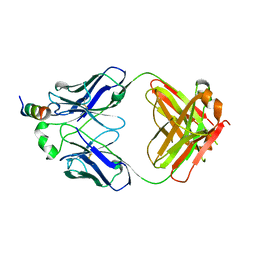 | |
7SKZ
 
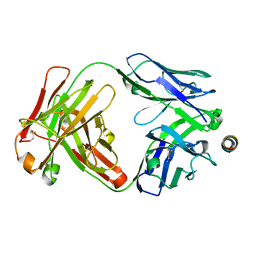 | |
6HKM
 
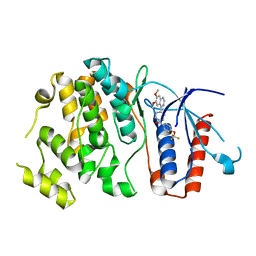 | | Crystal structure of Compound 1 with ERK5 | | Descriptor: | Mitogen-activated protein kinase 7, [4-(6,7-dimethoxyquinazolin-4-yl)piperidin-1-yl]-[4-(trifluoromethyloxy)phenyl]methanone | | Authors: | Nguyen, D, Lemos, C, Wortmann, L, Eis, K, Holton, S.J, Boemer, U, Lechner, C, Prechtl, S, Suelze, D, Siegel, F, Prinz, F, Lesche, R, Nicke, B, Mumberg, D, Bauser, M, Haegebarth, A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5.
J. Med. Chem., 62, 2019
|
|
8FY1
 
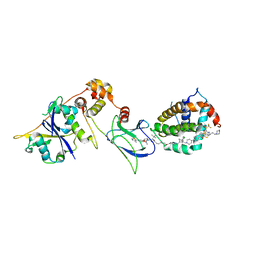 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY0
 
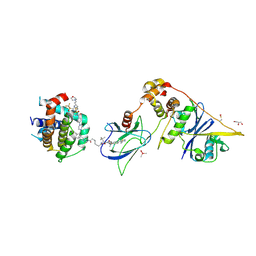 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
5MW6
 
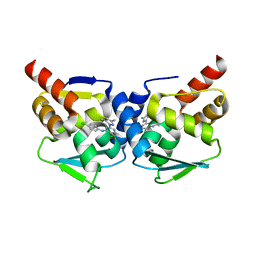 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
3AL1
 
 | | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOLAMINE, PROTEIN (D, ... | | Authors: | Patterson, W.R, Anderson, D.H, Degrado, W.F, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-26 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1.
Protein Sci., 8, 1999
|
|
2C8I
 
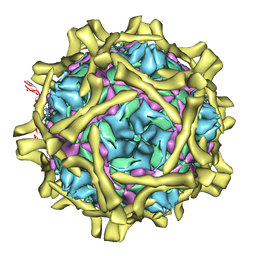 | | Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Pettigrew, D.M, Williams, D.T, Kerrigan, D, Evans, D.J, Lea, S.M, Bhella, D. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural and Functional Insights Into the Interaction of Echoviruses and Decay-Accelerating Factor.
J.Biol.Chem., 281, 2006
|
|
6TNH
 
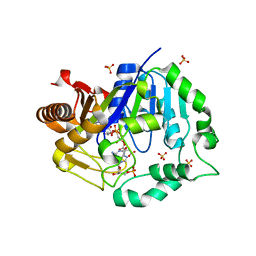 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6TNT
 
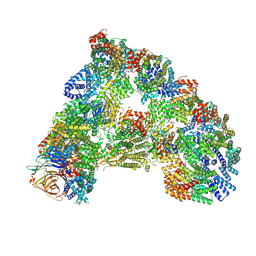 | | SUMOylated apoAPC/C with repositioned APC2 WHB domain | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Barford, D, Yatskevich, S. | | Deposit date: | 2019-12-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation.
Cell Rep, 34, 2021
|
|
6TFW
 
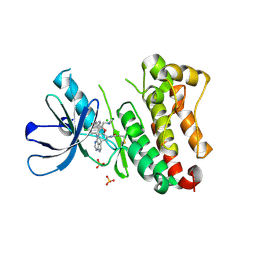 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 18d | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
6TGI
 
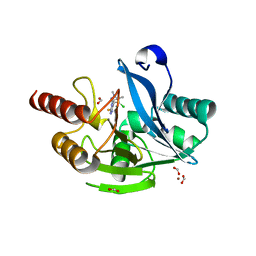 | | Crystal structure of VIM-2 in complex with triazole-based inhibitor OP24 | | Descriptor: | 5-(4-chloranyl-1,5-dimethyl-pyrazol-3-yl)-4-ethyl-1,2,4-triazole-3-thiol, FORMIC ACID, Vim-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
6TB9
 
 | | Capsid of native GTA particle computed with C5 symmetry | | Descriptor: | Head spike base Rcc01079, Head spike fiber Rcc01080, Major capsid protein Rcc01687 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
4U2W
 
 | | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin | | Descriptor: | 1,2-ETHANEDIOL, Bowman-Birk trypsin inhibitor, CALCIUM ION, ... | | Authors: | Grudnik, P, Golik, P, Malicki, S, Debowski, D, Legowska, A, Rolka, K, Dubin, G. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin.
Proteins, 83, 2015
|
|
6TA2
 
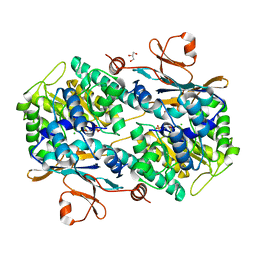 | | Human NAMPT in complex with nicotinic acid mononucleotide and phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6TD7
 
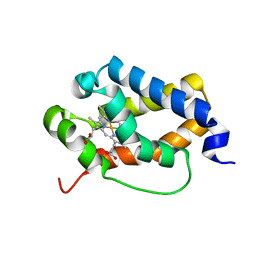 | | Structure of truncated hemoglobin THB11 from Chlamydomonas reinhardtii | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, THB11 | | Authors: | Huwald, D, Gasper, R, Hemschemeier, A, Hofmann, E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Distinctive structural properties of THB11, a pentacoordinate Chlamydomonas reinhardtii truncated hemoglobin with N- and C-terminal extensions.
J.Biol.Inorg.Chem., 25, 2020
|
|
6TH6
 
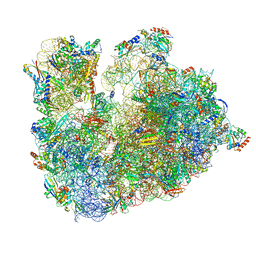 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-11-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
5Q06
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-(2-methyl-1,3-thiazol-4-yl)benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]sulfonylurea
To be published
|
|
4U4A
 
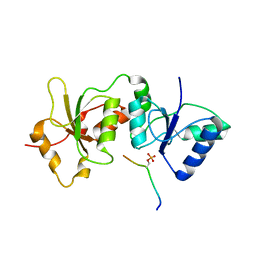 | |
