5O7B
 
 | |
1MXH
 
 | | Crystal Structure of Substrate Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | DIHYDROFOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
5ZS8
 
 | |
8WWC
 
 | |
1FZK
 
 | | MHC CLASS I NATURAL MUTANT H-2KBM1 HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudolph, M.G, Speir, J.A, Brunmark, A, Mattsson, N, Jackson, M.R, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-10-03 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity.
Immunity, 14, 2001
|
|
5OF0
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII), the E424M inactive mutant, in complex with a inhibitor CFBzOG | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[(4-fluorophenyl)methylamino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Motlova, L, Barinka, C. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
1MVF
 
 | | MazE addiction antidote | | Descriptor: | PemI-like protein 1, immunoglobulin heavy chain variable region | | Authors: | Loris, R, Marianovsky, I, Lah, J, Laeremans, T, Engelberg-Kulka, H, Glaser, G, Muyldermans, S, Wyns, L. | | Deposit date: | 2002-09-25 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the intrinsically flexible addiction antidote MazE.
J.Biol.Chem., 278, 2003
|
|
1MZZ
 
 | | Crystal Structure of Mutant (M182T)of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
1N0W
 
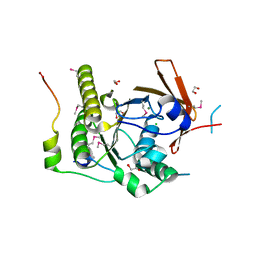 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
1N14
 
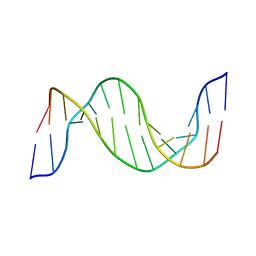 | | Structure and Dynamics of Thioguanine-modified Duplex DNA in Comparison with Unmodified DNA; Structure of Unmodified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*GP*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
1N2K
 
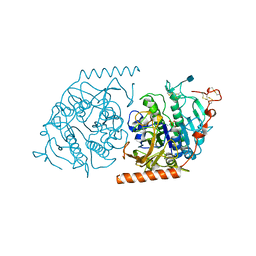 | | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, ... | | Authors: | Chruszcz, M, Laidler, P, Monkiewicz, M, Ortlund, E, Lebioda, L, Lewinski, K. | | Deposit date: | 2002-10-23 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A.
J.Inorg.Biochem., 96, 2003
|
|
4CRE
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-chloro-4-methyl-1H-quinolin-2-one, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
1N42
 
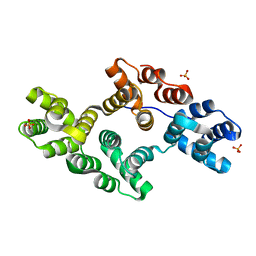 | | Crystal Structure of Annexin V R149E Mutant | | Descriptor: | Annexin V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interfacial basic cluster in annexin V couples phospholipid binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
4CPC
 
 | |
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
5X0W
 
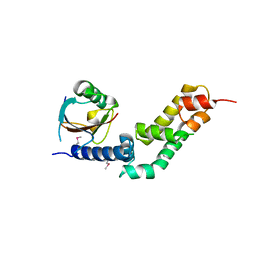 | | Molecular mechanism for the binding between Sharpin and HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Sharpin | | Authors: | Liu, J, Li, F, Cheng, X, Pan, L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly
Cell Rep, 21, 2017
|
|
5OLL
 
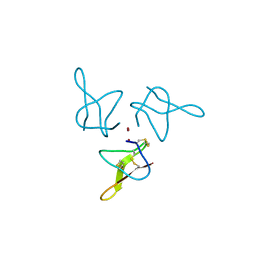 | | Crystal structure of gurmarin, a sweet taste suppressing polypeptide | | Descriptor: | Gurmarin, NICKEL (II) ION | | Authors: | Sigoillot, M, Neiers, F, Legrand, P, Roblin, P, Briand, L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Gurmarin, a Sweet Taste-Suppressing Protein: Identification of the Amino Acid Residues Essential for Inhibition.
Chem. Senses, 43, 2018
|
|
4CRG
 
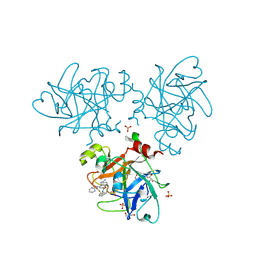 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-carbamimidoyl-N-phenyl-4-(pyrimidin-2-ylamino)naphthalene-2-carboxamide, COAGULATION FACTOR XI, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
5OEF
 
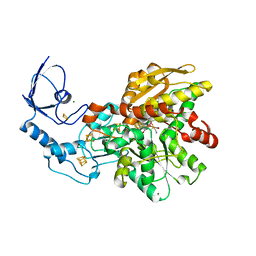 | | Active semisynthetic [FeFe]-hydrogenase CpI with aza-diselenato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Kertess, L, Esselborn, J, Happe, T, Hofmann, E. | | Deposit date: | 2017-07-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chalcogenide substitution in the [2Fe] cluster of [FeFe]-hydrogenases conserves high enzymatic activity.
Dalton Trans, 46, 2017
|
|
4CR9
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 4-methylquinoline-2,6-diamine, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CTE
 
 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S in complex with a thio-oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, 1-thio-beta-D-glucopyranose-(1-3)-1-thio-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CRA
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | COAGULATION FACTOR XI, GLYCEROL, N-[(1S)-2-[(2-amino-5-quinolyl)methylamino]-1-benzyl-2-oxo-ethyl]-4-hydroxy-2-oxo-1H-quinoline-6-carboxamide, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CRD
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | COAGULATION FACTOR XI, Methyl N-[4-[5-chloro-2-[[3-[5-chloro-2-(tetrazol-1-yl)phenyl]propanoylamino]methyl]-1H-imidazol-4-yl]phenyl]carbamate, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
5WLX
 
 | |
1MSZ
 
 | | Solution structure of the R3H domain from human Smubp-2 | | Descriptor: | DNA-binding protein SMUBP-2 | | Authors: | Liepinsh, E, Leonchiks, A, Sharipo, A, Guignard, L, Otting, G. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the R3H domain from human Smubp-2
J.Mol.Biol., 326, 2003
|
|
