5XC3
 
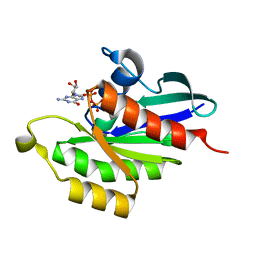 | |
5UYZ
 
 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) mutant His147Arg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
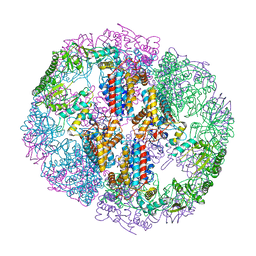
5UYX
 
 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
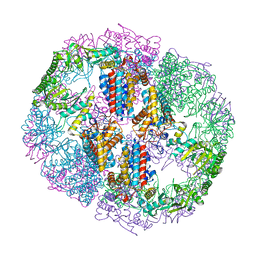
5VTB
 
 | | Crystal structure of RBBP4 bound to BCL11a peptide | | Descriptor: | B-cell lymphoma/leukemia 11A, GLYCEROL, Histone-binding protein RBBP4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-05-16 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes.
J. Biol. Chem., 293, 2018
|
|
5VJA
 
 | | Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) | | Descriptor: | (2R)-2-{[1-(3-chlorophenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]sulfanyl}propanamide, DIMETHYL SULFOXIDE, Death-associated protein kinase 3, ... | | Authors: | Carlson, D.A, Singer, M.R, Sutherland, C, Redondo, C, Alexander, L, Hughes, P.F, Knapp, S, MacDonald, J.A, Haystead, T.A.J. | | Deposit date: | 2017-04-19 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Targeting Pim Kinases and DAPK3 to Control Hypertension.
Cell Chem Biol, 25, 2018
|
|
2HPD
 
 | | CRYSTAL STRUCTURE OF HEMOPROTEIN DOMAIN OF P450BM-3, A PROTOTYPE FOR MICROSOMAL P450'S | | Descriptor: | CYTOCHROME P450 BM-3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ravichandran, K.G, Boddupalli, S.S, Hasemann, C.A, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-09-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's.
Science, 261, 1993
|
|
3CHM
 
 | |
1QAT
 
 | |
1QOM
 
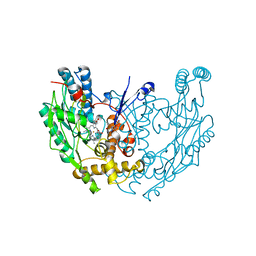 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH SWAPPED N-TERMINAL HOOK | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Rosenfeld, R.A, Arvai, A.S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-15 | | Release date: | 1999-12-15 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-Terminal Domain Swapping and Metal Ion Binding in Nitric Oxide Synthase Dimerization
Embo J., 18, 1999
|
|
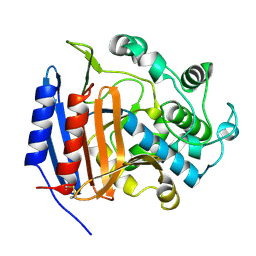
3PTE
 
 | |
1RKA
 
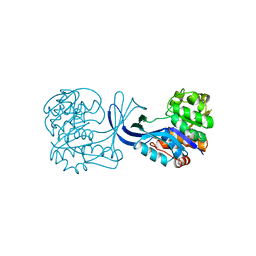 | |
2QPE
 
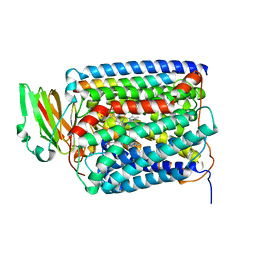 | | An unexpected outcome of surface-engineering an integral membrane protein: Improved crystallization of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Luna, V.M, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba(3) from Thermus thermophilus.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1PW8
 
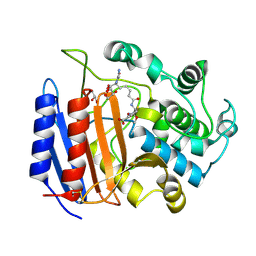 | | Covalent Acyl Enzyme Complex Of The R61 DD-Peptidase with A Highly Specific Cephalosporin | | Descriptor: | (6R,7R)-3-[(ACETYLOXY)METHYL]-7-{[(6S)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-8-OXO-5-THIA-1-AZABICYCLO[4.2.0]OCTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PWC
 
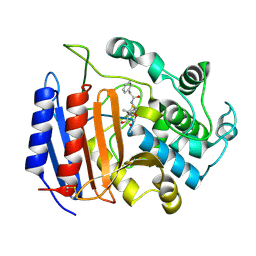 | | penicilloyl acyl enzyme complex of the Streptomyces R61 DD-peptidase with penicillin G | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, OPEN FORM - PENICILLIN G | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1R8O
 
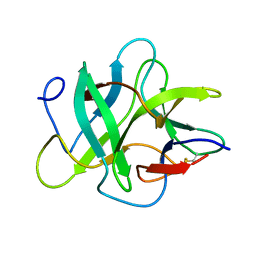 | | Crystal structure of an unusual Kunitz-type trypsin inhibitor from Copaifera langsdorffii seeds | | Descriptor: | Kunitz trypsin inhibitor | | Authors: | Krauchenco, S, Nagem, R.A.P, da Silva, J.A, Marangoni, S, Polikarpov, I. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Biochimie, 86, 2004
|
|
3QS2
 
 | |
2SSP
 
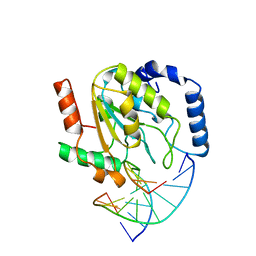 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
2PIL
 
 | | Crystallographic Structure of Phosphorylated Pilin from Neisseria: Phosphoserine Sites Modify Type IV Pilus Surface Chemistry | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Dunham, S.A, Koomey, M, Tainer, J.A. | | Deposit date: | 1998-03-02 | | Release date: | 1998-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic structure reveals phosphorylated pilin from Neisseria: phosphoserine sites modify type IV pilus surface chemistry and fibre morphology.
Mol.Microbiol., 31, 1999
|
|
3QS3
 
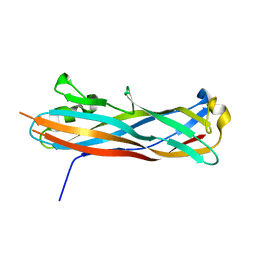 | |
2SAS
 
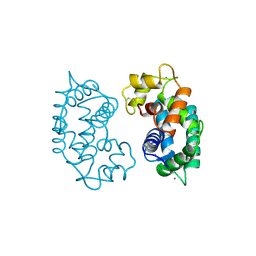 | |
1PWG
 
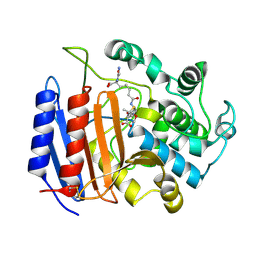 | | Covalent Penicilloyl Acyl Enzyme Complex Of The Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(6S)-6-carboxy-6-(glycylamino)hexanoyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.074 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
3RCZ
 
 | | Rad60 SLD2 Ubc9 Complex | | Descriptor: | DNA repair protein rad60, SUMO-conjugating enzyme ubc9 | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
2OLP
 
 | | Structure and ligand selection of hemoglobin II from Lucina pectinata | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gavira, J.A, Camara-Artigas, A, de Jesus, W, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2007-01-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
J.Biol.Chem., 283, 2008
|
|
3N37
 
 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase 2 subunit beta | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
3N38
 
 | | Ribonucleotide Reductase NrdF from Escherichia coli Soaked with Ferrous Ions | | Descriptor: | FE (II) ION, Ribonucleoside-diphosphate reductase 2 subunit beta | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
