6NHW
 
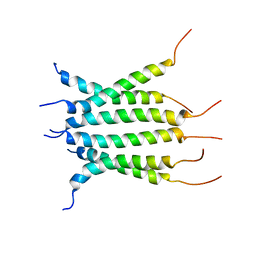 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6DMD
 
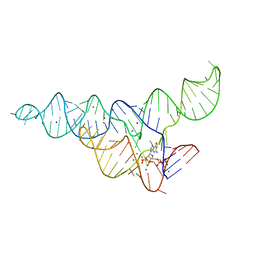 | | ppGpp Riboswitch bound to ppGpp, manganese chloride structure | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Peselis, A, Serganov, A. | | Deposit date: | 2018-06-05 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands.
Nat. Chem. Biol., 14, 2018
|
|
6NHY
 
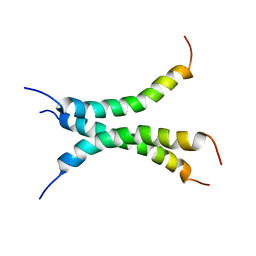 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6DNR
 
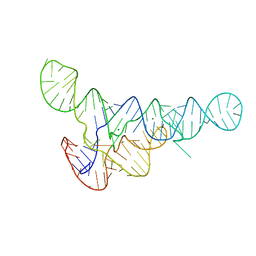 | |
3UI4
 
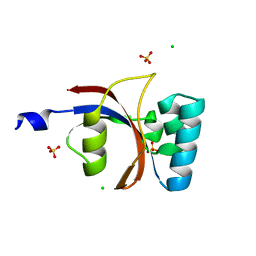 | | 0.8 A resolution crystal structure of human Parvulin 14 | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SULFATE ION | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
4OEP
 
 | |
6NUG
 
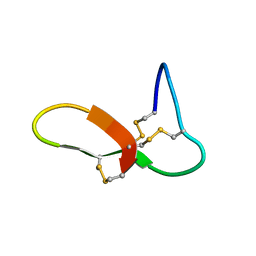 | | hGRNA4-28_3s | | Descriptor: | Granulin-4 | | Authors: | Dastpeyman, M, Bansal, P, Wilson, D, Loukas, A, Smout, M, Daly, N. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | hGRN4-28_3s
to be published
|
|
6UAN
 
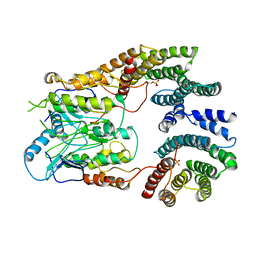 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
1MOR
 
 | |
6HXA
 
 | | AntI from P. luminescens catalyses terminal polyketide shortening in the biosynthesis of anthraquinones | | Descriptor: | AntI, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Zhou, Q, Braeuer, A, Grammbitter, G, Schmalhofer, M, Saura, P, Adihou, H, Kaila, V.R.I, Groll, M, Bode, H. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of polyketide shortening in anthraquinone biosynthesis ofPhotorhabdus luminescens.
Chem Sci, 10, 2019
|
|
2R8B
 
 | | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | SULFATE ION, Uncharacterized protein Atu2452 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-10 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
6O6O
 
 | | Structure of the regulator FasR from Mycobacterium tuberculosis | | Descriptor: | MYRISTIC ACID, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6O5V
 
 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
2R9N
 
 | | Cathepsin S complexed with Compound 26 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(3S)-1-benzyl-3-cyanopyrrolidin-3-yl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
2R9O
 
 | | Cathepsin S complexed with Compound 8 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
3UI5
 
 | | Crystal structure of human Parvulin 14 | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
1M4L
 
 | | STRUCTURE OF NATIVE CARBOXYPEPTIDASE A AT 1.25 RESOLUTION | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Kilshtain-Vardi, A, Glick, M, Greenblatt, H.M, Goldblum, A, Shoham, G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Refined structure of bovine carboxypeptidase A at 1.25 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6O81
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B bound to translation initiation factor 2 from Homo sapiens | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Nguyen, H, Kenner, L, Frost, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.
Science, 364, 2019
|
|
2RD4
 
 | | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 1, Phospholipase A2 isoform 2, ... | | Authors: | Mirza, Z, Kaur, A, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution
To be Published
|
|
4KIV
 
 | |
6O6D
 
 | |
5HZ6
 
 | |
1MQP
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
6DY1
 
 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) with fatty acid (myristate), in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, CHLORIDE ION, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4OQ1
 
 | |
