4EZH
 
 | |
7W8L
 
 | | Crystal Structure of Co-type nitrile hydratase mutant from Pseudonocardia thermophila - M46R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
7W8M
 
 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
4A3N
 
 | | Crystal Structure of HMG-BOX of Human SOX17 | | Descriptor: | TRANSCRIPTION FACTOR SOX-17, ZINC ION | | Authors: | Gao, N, Gao, H, Qian, H, Si, S, Xie, Y. | | Deposit date: | 2011-10-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Human Transcription Factor Sry-Related Box 17 Binding to DNA.
Protein Pept.Lett., 20, 2013
|
|
2XUE
 
 | | CRYSTAL STRUCTURE OF JMJD3 | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Rowland, P, Mosley, J, Thomas, P.J. | | Deposit date: | 2010-10-19 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
4ASK
 
 | | CRYSTAL STRUCTURE OF JMJD3 WITH GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, COBALT (II) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Mosley, J, Liddle, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
4NA5
 
 | |
4NA0
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4N9Y
 
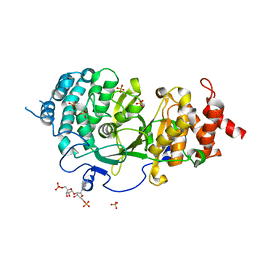 | |
4N9Z
 
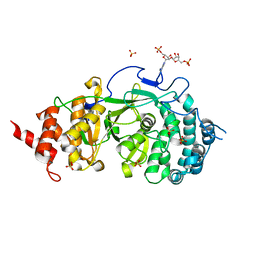 | |
4NA6
 
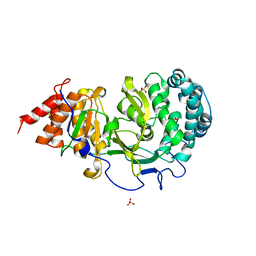 | |
4FC2
 
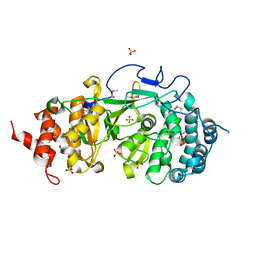 | |
8JFU
 
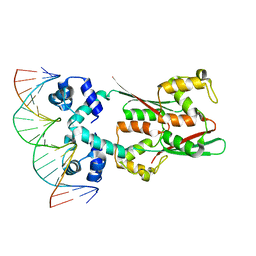 | | AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFO
 
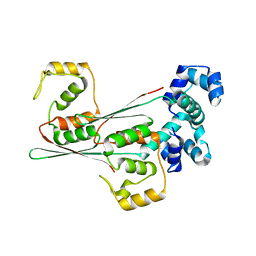 | |
8JFT
 
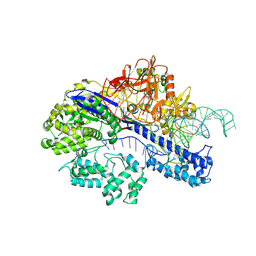 | | Cryo-EM structure of SaCas9-AcrIIA15 CTD-sgRNA complex | | Descriptor: | AcrIIA15, CRISPR-associated endonuclease Cas9, sgRNA of SaCas9 | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFR
 
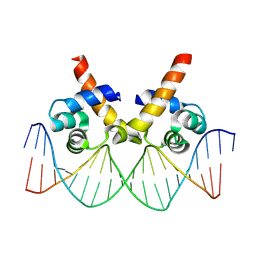 | | N-terminal domain of AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JG9
 
 | |
6N05
 
 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
7BV7
 
 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
7XVI
 
 | |
7XVK
 
 | |
5WIH
 
 | |
5GNF
 
 | | Crystal structure of anti-CRISPR protein AcrF3 | | Descriptor: | CALCIUM ION, Uncharacterized protein AcrF3 | | Authors: | Wang, J, Wang, Y. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
3TJP
 
 | | Crystal Structure of PI3K gamma with N6-(3,4-dimethoxyphenyl)-2-morpholino-[4,5'-bipyrimidine]-2',6-diamine | | Descriptor: | N~6~-(3,4-dimethoxyphenyl)-2-(morpholin-4-yl)-4,5'-bipyrimidine-2',6-diamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, R.A, Ornelas, E. | | Deposit date: | 2011-08-24 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Identification of 5-(2,4-dimorpholinopyrimidin-6-yl)-4-(trifluoromethyl)pyridin-2-amine (NVP-BKM120) as a Potent, Selective and Orally Bioavailable Class I PI3 Kinase Inhibitor for the Treatment of Cancer
To be Published
|
|
