4D28
 
 | |
4U36
 
 | | Crystal structure of a seed lectin from Vatairea macrocarpa complexed with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
1QHE
 
 | |
4AB4
 
 | | Structure of Xenobiotic Reductase B from Pseudomonas putida in complex with TNT | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-TRINITROTOLUENE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Carvalho, A.L, Mukhopadhyay, A, Romao, M.J, Bursakov, S, Kladova, A, Ramos, J.L, van Dillewijn, P. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants of Aromatic Ring Reduction in the Xenobiotic Reductase B from Pseudomonas Putida
To be Published
|
|
1CAV
 
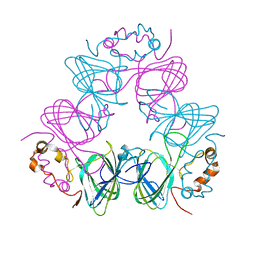 | |
2ESH
 
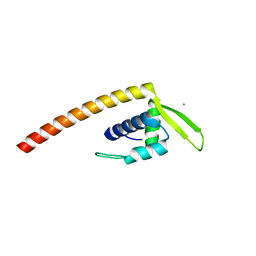 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
3TXM
 
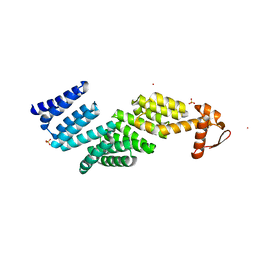 | |
2PMO
 
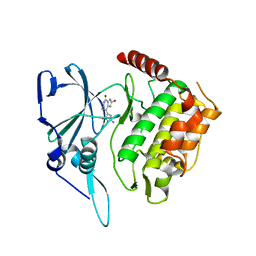 | | Crystal structure of PfPK7 in complex with hymenialdisine | | Descriptor: | 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, Ser/Thr protein kinase | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
2H3X
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes Faecalis (Form 3) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
6U83
 
 | | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-ALANINE, Outer membrane associated protein, ... | | Authors: | Michalska, K, Skarina, T, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3566 Å) | | Cite: | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
2POY
 
 | | Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 in complex with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryptosporidium Parvum Cyclophilin Type Peptidyl-Prolyl Cis-Trans Isomerase Cgd2_4120 in Complex with Cyclosporin A.
To be Published
|
|
6UIU
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | N-(2-{bis[(pyridin-2-yl)methyl]amino}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide, Streptavidin | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
1V6N
 
 | | Peanut lectin with 9mer peptide (PVIWSSATG) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4U4I
 
 | | Megavirus chilensis superoxide dismutase | | Descriptor: | Cu/Zn superoxide dismutase | | Authors: | Lartigue, A, Claverie, J.-M, Burlat, B, Coutard, B, Abergel, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The megavirus chilensis cu,zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme.
J.Virol., 89, 2015
|
|
4BCA
 
 | | MAMMALIAN ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE: Tyr578Phe mutant | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, CHLORIDE ION, ... | | Authors: | Nenci, S, Piano, V, Rosati, S, Aliverti, A, Pandini, V, Fraaije, M.W, Heck, A.J.R, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precursor of Ether Phospholipids is Synthesized by a Flavoenzyme Through Covalent Catalysis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F9P
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 103.2 Single Chain Antibody | | Descriptor: | 103.2 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.519 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
5VF6
 
 | |
1DEG
 
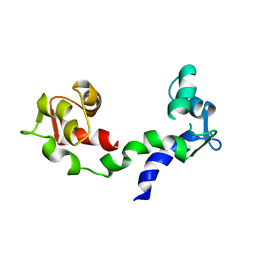 | | THE LINKER OF DES-GLU84 CALMODULIN IS BENT AS SEEN IN THE CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Raghunathan, S, Chandross, R, Cheng, B.P, Persechini, A, Sobottk, S.E, Kretsinger, R.H. | | Deposit date: | 1993-06-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The linker of des-Glu84-calmodulin is bent.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
2BC9
 
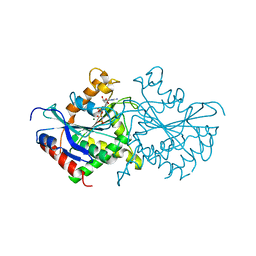 | | Crystal-structure of the N-terminal large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with non-hydrolysable GTP analogue GppNHp | | Descriptor: | Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
4AT3
 
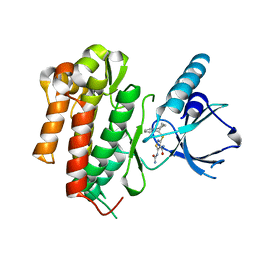 | | CRYSTAL STRUCTURE OF TRKB KINASE DOMAIN IN COMPLEX WITH CPD5N | | Descriptor: | (5Z)-5-(carbamoylimino)-3-[(5R)-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ylsulfanyl]-2,5-dihydroisothiazole-4-carboxamide, BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4B9X
 
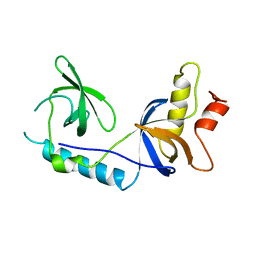 | | Structure of extended Tudor domain TD3 from mouse TDRD1 | | Descriptor: | TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Multiple Tudor Domain-Containing Protein Tdrd1 is a Molecular Scaffold for Mouse Piwi Proteins and Pirna Biogenesis Factors.
RNA, 18, 2012
|
|
4FFL
 
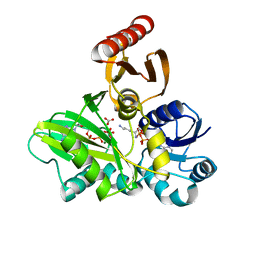 | | PylC in complex with L-lysine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
4AVD
 
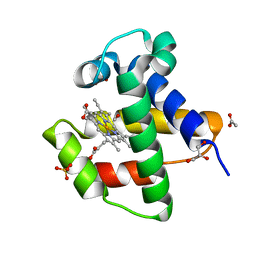 | | C.lacteus nerve Hb in complex with CO | | Descriptor: | ACETATE ION, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Germani, F, Pesce, A, Venturini, A, Moens, L, Bolognesi, M, Dewilde, S, Nardini, M. | | Deposit date: | 2012-05-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structures of the Cerebratulus Lacteus Mini-Hb in the Unligated and Carbomonoxy States.
Int.J.Mol.Sci., 13, 2012
|
|
4AVX
 
 | | Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) bound to an IP2 compound at 1.68 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ... | | Authors: | Williams, E, Canning, P, Shrestha, L, Krojer, T, Vollmar, M, Slowey, A, Conway, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Tandem Vhs and Fyve Domains of Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) Bound to an Ip2 Compound at 1.68 A Resolution
To be Published
|
|
4FR4
 
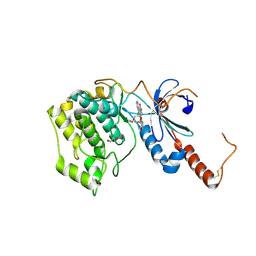 | | Crystal structure of human serine/threonine-protein kinase 32A (YANK1) | | Descriptor: | 1,2-ETHANEDIOL, STAUROSPORINE, Serine/threonine-protein kinase 32A | | Authors: | Chaikuad, A, Elkins, J.M, Krojer, T, Mahajan, P, Goubin, S, Szklarz, M, Tumber, A, Wang, J, Savitsky, P, Shrestha, B, Daga, N, Picaud, S, Fedorov, O, Allerston, C.K, Latwiel, S.V.A, Vollmar, M, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human serine/threonine-protein kinase 32A (YANK1)
To be Published
|
|
