7LIQ
 
 | |
7LIP
 
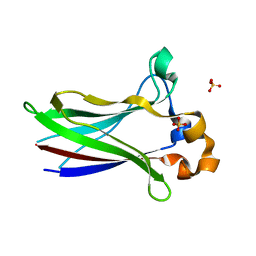 | | X-ray structure of SPOP MATH domain (D140G) | | Descriptor: | SULFATE ION, Speckle-type POZ protein | | Authors: | Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.
Sci Adv, 7, 2021
|
|
5W9J
 
 | |
5W9P
 
 | |
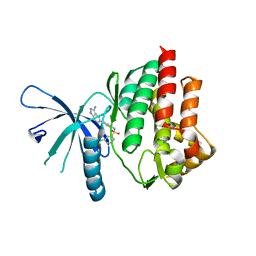
4E20
 
 | |
7E8I
 
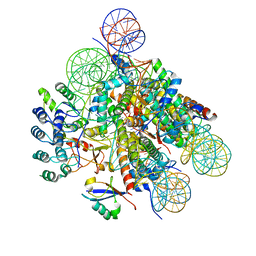 | | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (145-MER), Histone H2A, ... | | Authors: | Dai, Y, Dai, L, Zhou, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin.
Mol.Cell, 81, 2021
|
|
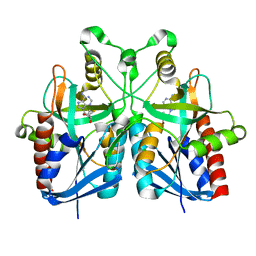
5B7P
 
 | |
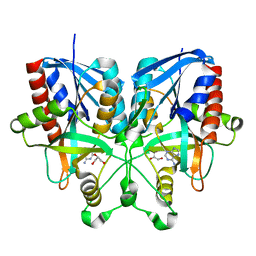
5B7G
 
 | |
5W9H
 
 | |
5C0S
 
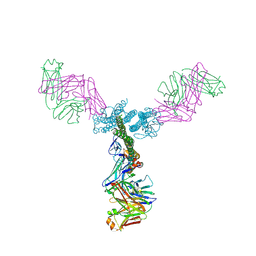 | | Crystal structure of a generation 4 influenza hemagglutinin stabilized stem in complex with the broadly neutralizing antibody CR6261 | | Descriptor: | CR6261 antibody heavy chain, CR6261 antibody light chain, Hemagglutinin, ... | | Authors: | Boyington, J.C, Kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
5W9K
 
 | |
3KA0
 
 | |
3KC3
 
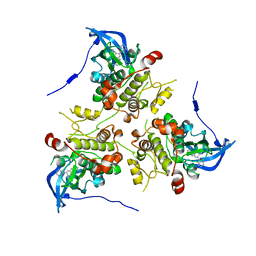 | | MK2 complexed to inhibitor N4-(7-(benzofuran-2-yl)-1H-indazol-5-yl)pyrimidine-2,4-diamine | | Descriptor: | MAP kinase-activated protein kinase 2, N~4~-[7-(1-benzofuran-2-yl)-1H-indazol-5-yl]pyrimidine-2,4-diamine | | Authors: | Argiriadi, M.A, Talanian, R.V, Borhani, D.W. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2,4-Diaminopyrimidine MK2 inhibitors. Part I: Observation of an unexpected inhibitor binding mode.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5B7N
 
 | |
5W9O
 
 | |
5W9L
 
 | |
6J0Q
 
 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
3HL5
 
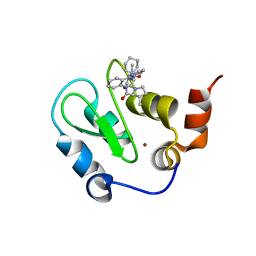 | | Crystal structure of XIAP BIR3 with CS3 | | Descriptor: | (3S)-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-3-methyl-N-(2-pyrimidin-2-ylphenyl)-L-prolinamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism of c-IAP and XIAP proteins is required for efficient induction of cell death by small-molecule IAP antagonists.
Acs Chem.Biol., 4, 2009
|
|
4E1Z
 
 | |
6A6J
 
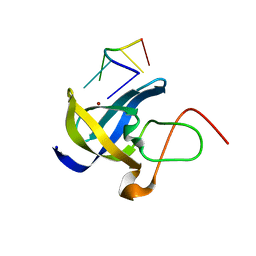 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
6KP6
 
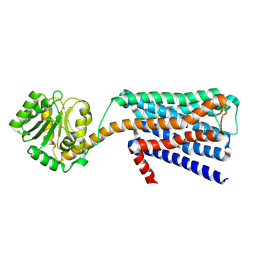 | | The structural study of mutation induced inactivation of Human muscarinic receptor M4 | | Descriptor: | Muscarinic acetylcholine receptor M4,GlgA glycogen synthase,Muscarinic acetylcholine receptor M4 | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural study of mutation-induced inactivation of human muscarinic receptor M4
Iucrj, 7, 2020
|
|
3GT9
 
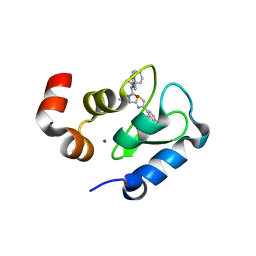 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | Baculoviral IAP repeat-containing 7, N-{(1S)-1-cyclohexyl-2-[(2S)-2-(4-naphthalen-1-yl-1,3-thiazol-2-yl)pyrrolidin-1-yl]-2-oxoethyl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6IH4
 
 | |
6A82
 
 | |
6AGL
 
 | |
