5C1U
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound Xb | | Descriptor: | (2S)-2-[[(E)-3-[4-(dimethylamino)phenyl]prop-2-enoyl]amino]-N-[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-phenyl-propanamide, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1X
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound VIII | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1Y
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound 1 | | Descriptor: | 3C proteinase, propan-2-yl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
1YT9
 
 | | HIV Protease with oximinoarylsulfonamide bound | | Descriptor: | (S)-N-((2S,3R)-3-HYDROXY-4-(4-((E)-(HYDROXYIMINO)METHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YL)-3-METHYL-2-(3 -((2-METHYLTHIAZOL-4-YL)METHYL)-2-OXOIMIDAZOLIDIN-1-YL)BUTANAMIDE, Pol polyprotein | | Authors: | Yeung, C.M, Klein, L.L, Flentge, C.A, Randolph, J.T, Zhao, C, Sun, M, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | Deposit date: | 2005-02-10 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oximinoarylsulfonamides as potent HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3MTV
 
 | | The Crystal Structure of the PRRSV Nonstructural Protein Nsp1 | | Descriptor: | Papain-like cysteine protease | | Authors: | Xue, F, Sun, Y.N, Yan, L.M, Zhao, C, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease
J.Virol., 84, 2010
|
|
8SAI
 
 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5YKJ
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | GLYCEROL, Peroxiredoxin PRX1, mitochondrial, ... | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-14 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
To be published
|
|
5YKW
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | Thioredoxin-3, mitochondrial, peptide THR-PRO-VAL-CYS-THR-THR-GLU-VAL | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
to be published
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
7VH5
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
8JLK
 
 | | Ulotaront(SEP-363856)-bound mTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
5ZMR
 
 | | Solution Structure of the N-terminal Domain of the Yeast Rpn5 | | Descriptor: | 26S proteasome regulatory subunit RPN5 | | Authors: | Zhang, W, Zhao, C, Li, H, Hu, Y, Jin, C. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of proteasome lid subunit Rpn5
Biochem. Biophys. Res. Commun., 504, 2018
|
|
8JLQ
 
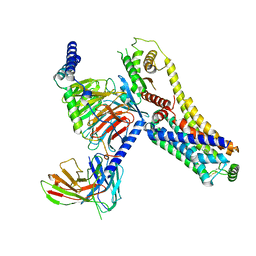 | | Fenoldopam-bound hTAAR1-Gs protein complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLN
 
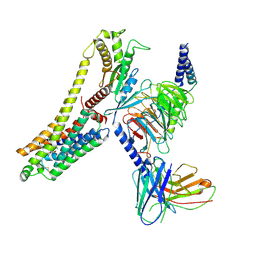 | | T1AM-bound hTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLP
 
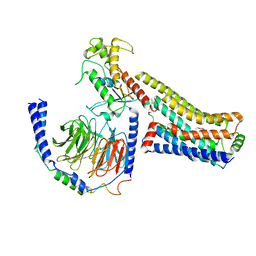 | | Ralmitaront(RO-6889450)-bound hTAAR1-Gs protein complex | | Descriptor: | 5-ethyl-4-methyl-~{N}-[4-[(2~{S})-morpholin-2-yl]phenyl]-1~{H}-pyrazole-3-carboxamide, Gs, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLO
 
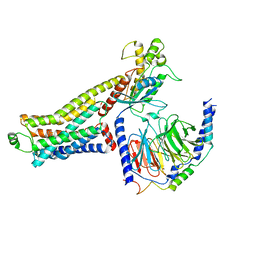 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLR
 
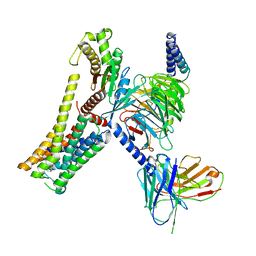 | | A77636-bound hTAAR1-Gs protein complex | | Descriptor: | (1~{S},3~{R})-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1~{H}-isochromene-5,6-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSP
 
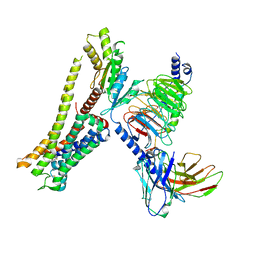 | | Ulotaront(SEP-363856)-bound Serotonin 1A (5-HT1A) receptor-Gi complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, 5-hydroxytryptamine receptor 1A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLJ
 
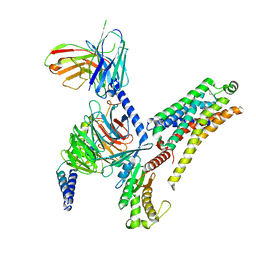 | | T1AM-bound mTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSO
 
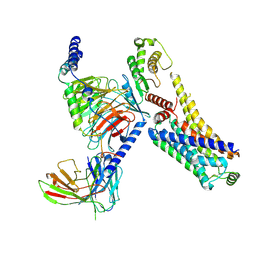 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
2MX4
 
 | | NMR structure of Phosphorylated 4E-BP2 | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 2 | | Authors: | Bah, A, Forman-Kay, J, Vernon, R, Siddiqui, Z, Krzeminski, M, Muhandiram, R, Zhao, C, Sonenberg, N, Kay, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch.
Nature, 519, 2015
|
|
7BOR
 
 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7CRD
 
 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
