3BCG
 
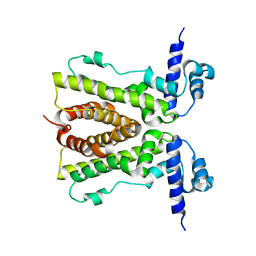 | | Conformational changes of the AcrR regulator reveal a mechanism of induction | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Gu, R, Li, M, Su, C.C, Long, F, Yang, F, McDermott, G, Yu, E.Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational change of the AcrR regulator reveals a possible mechanism of induction.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
9BFM
 
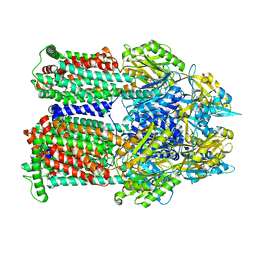 | | Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor | | Descriptor: | (2S)-1-(3,4-dichlorophenoxy)-3-(4-{[4-(trifluoromethyl)pyrimidin-2-yl]amino}piperidin-1-yl)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFH
 
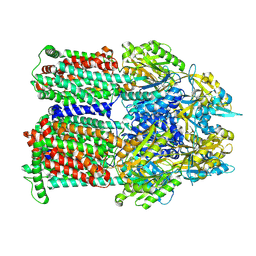 | | Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor | | Descriptor: | (2S)-1-[(3R)-3-aminopyrrolidin-1-yl]-3-(3,4-dichlorophenoxy)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFT
 
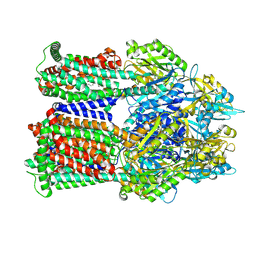 | | Cryo-EM co-structure of AcrB with CU244 | | Descriptor: | (2S)-1-{[(1R,5R)-3-azabicyclo[3.1.0]hexan-6-yl]amino}-3-(3,5-dichlorophenoxy)propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFN
 
 | | Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor | | Descriptor: | (2R)-1-(4-aminopiperidin-1-yl)-3-[3-(trifluoromethyl)phenoxy]propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
7M4P
 
 | | Multidrug Efflux pump AdeJ with Eravacycline bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Efflux pump membrane transporter, Eravacycline | | Authors: | Zhang, Z. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4Q
 
 | | Multidrug Efflux pump AdeJ | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
8DNP
 
 | | Human Brain Ferritin Heavy Chain | | Descriptor: | FE (III) ION, Ferritin heavy chain | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
8DNU
 
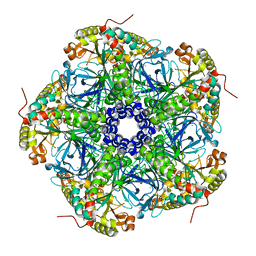 | | Human Brain Glutamine Synthetase | | Descriptor: | Glutamine synthetase, MANGANESE (II) ION | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
8DNO
 
 | |
8DNS
 
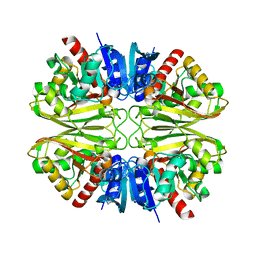 | | Human Brain Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
8DNM
 
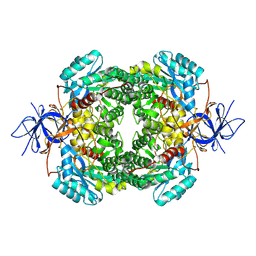 | | Human Brain Dihydropyrimidinase-related protein 2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
6BHP
 
 | |
4NN1
 
 | |
7SFL
 
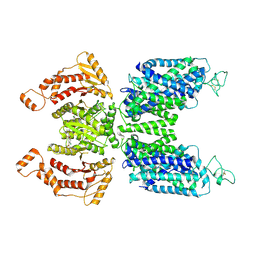 | | Human NKCC1 state Fu-II | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
5D1R
 
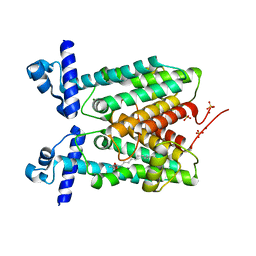 | | Crystal structure of Mycobacterium tuberculosis Rv1816 transcriptional regulator. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Rv1816 transcriptional regulator, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
5D18
 
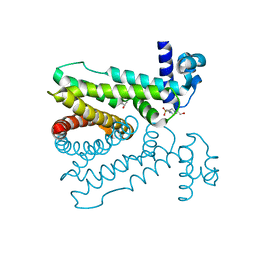 | | Crystal structure of Mycobacterium tuberculosis Rv0302, form I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis transcriptional regulator Rv0302.
Protein Sci., 2015
|
|
7SMP
 
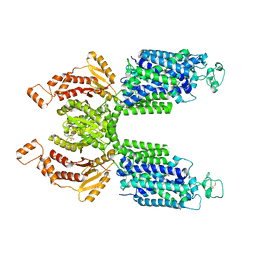 | | CryoEM structure of NKCC1 Bu-I | | Descriptor: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
5D9R
 
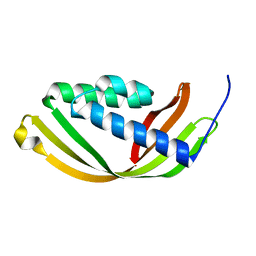 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | Descriptor: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | Authors: | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
5D19
 
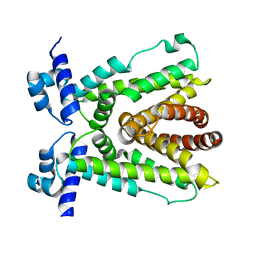 | |
5D1W
 
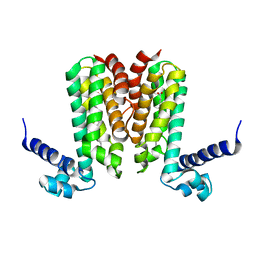 | | Crystal structure of Mycobacterium tuberculosis Rv3249c transcriptional regulator. | | Descriptor: | PALMITIC ACID, Rv3249c transcriptional regulator | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4K34
 
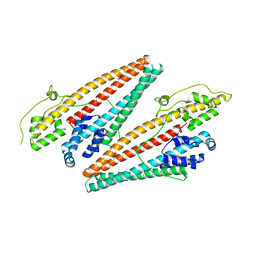 | |
4K7K
 
 | |
5N8P
 
 | |
5N97
 
 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
