5OOO
 
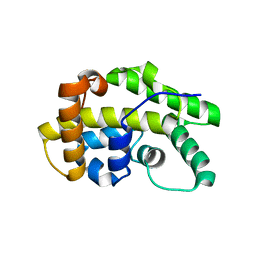 | |
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
6ADH
 
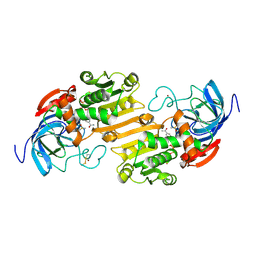 | |
7ADH
 
 | |
9ENG
 
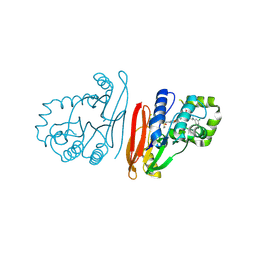 | | Structure of K.pneumoniae LpxH in complex with EBL-3218 | | Descriptor: | MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase, ~{N}-[4-[4-[3,5-bis(chloranyl)phenyl]piperazin-1-yl]sulfonylphenyl]-3-(methylsulfonylamino)benzamide | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and in vitro biological evaluation of meta-sulfonamidobenzamide-based antibacterial LpxH inhibitors.
Eur.J.Med.Chem., 278, 2024
|
|
4WCD
 
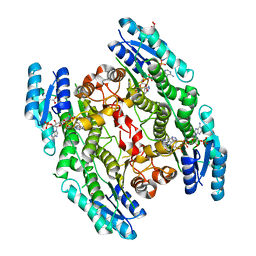 | | Trypanosoma brucei PTR1 in complex with inhibitor 10 | | Descriptor: | 5-(1H-benzotriazol-6-yl)-1,3,4-thiadiazol-2-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
8ADH
 
 | |
5JCX
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor NP-29 | | Descriptor: | 3,5,7-trihydroxy-2-(2-hydroxyphenyl)-4H-1-benzopyran-4-one, ACETATE ION, GLYCEROL, ... | | Authors: | Landi, G, Pozzi, C, Di Pisa, F, Dello Iacono, L, Mangani, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Profiling of Flavonol Derivatives for the Development of Antitrypanosomatidic Drugs.
J.Med.Chem., 59, 2016
|
|
5JDI
 
 | | Trypanosoma brucei PTR1 in complex with cofactor and inhibitor NMT-H024 (compound 2) | | Descriptor: | 3,6-dihydroxy-2-(3-hydroxyphenyl)-4H-1-benzopyran-4-one, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Landi, G, Pozzi, C, Di Pisa, F, Dello lacono, L, Mangani, S. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Profiling of Flavonol Derivatives for the Development of Antitrypanosomatidic Drugs.
J.Med.Chem., 59, 2016
|
|
5NNS
 
 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
5ADH
 
 | |
6YE4
 
 | | Structure of ExbB pentamer from Serratia marcescens by single particle cryo electron microscopy | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Biopolymer transport protein ExbB | | Authors: | Biou, V, Delepelaire, P, Coureux, P.D, Chami, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
7AJQ
 
 | | cryo-EM structure of ExbBD from Serratia Marcescens | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD | | Authors: | Biou, V, Adaixo, R, Coureux, P.D, Delepelaire, P, Chami, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
5L42
 
 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 3 | | Descriptor: | (2~{R})-2-[3,4-bis(oxidanyl)phenyl]-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
5L4N
 
 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 1 | | Descriptor: | (2~{R})-2-(3-hydroxyphenyl)-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
5K6A
 
 | | Trypanosoma brucei Pteridine reductase 1 (PTR1) in complex with compound 1 | | Descriptor: | (2~{R})-2-(3-hydroxyphenyl)-6-oxidanyl-2,3-dihydrochromen-4-one, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Landi, G, Pozzi, C, Di Pisa, F, Dello lacono, L, Mangani, S. | | Deposit date: | 2016-05-24 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
7TTZ
 
 | | Heterodimeric IgA Fc in complex with Staphylococcus aureus protein SSL7 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boulanger, M.J, Verstraete, M, Heinkel, F, Escobar, E, Dixit, S, Von Kreudenstein, T.S. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a pure and stable heterodimeric IgA for the development of multispecific therapeutics.
Mabs, 14, 2022
|
|
7NBT
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 21 | | Descriptor: | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NEO
 
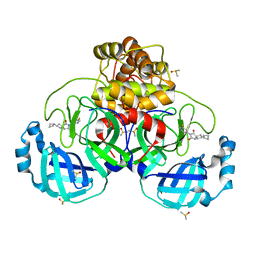 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 15 | | Descriptor: | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7AU4
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 3 | | Descriptor: | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B77
 
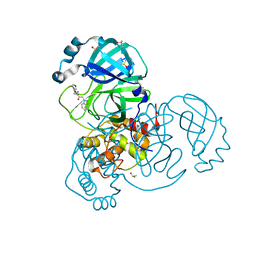 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 8 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-ethyl-~{N}-(furan-3-ylmethyl)ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B2J
 
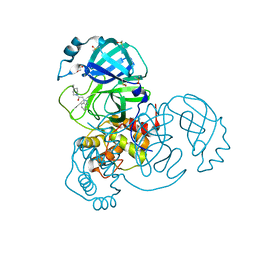 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 5 | | Descriptor: | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B2U
 
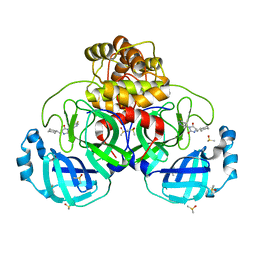 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 1 | | Descriptor: | (5S)-5-(cyclohexylmethyl)-3-(5-fluoropyridin-3-yl)imidazolidine-2,4-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B5Z
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 6 | | Descriptor: | 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-methylenepiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7QBB
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 18 | | Descriptor: | 3C-like proteinase nsp5, 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
