3PXZ
 
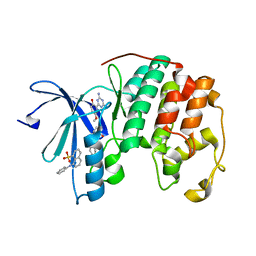 | | CDK2 ternary complex with JWS648 and ANS | | Descriptor: | 2-(4,6-diamino-1,3,5-triazin-2-yl)-4-methoxyphenol, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
7THT
 
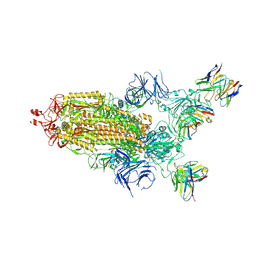 | | CryoEM structure of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1042 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 heavy chain, ... | | Authors: | Manne, K, May, A, Acharya, P. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
7TL9
 
 | |
3PXF
 
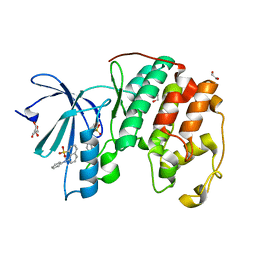 | | CDK2 in complex with two molecules of 8-anilino-1-naphthalene sulfonate | | Descriptor: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
7TL1
 
 | |
3PXR
 
 | | Apo CDK2 crystallized from Jeffamine | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PY0
 
 | | CDK2 in complex with inhibitor SU9516 | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, 1,2-ETHANEDIOL, Cell division protein kinase 2, ... | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PXY
 
 | | CDK2 in complex with inhibitor JWS648 | | Descriptor: | 2-(4,6-diamino-1,3,5-triazin-2-yl)-4-methoxyphenol, Cell division protein kinase 2, PHOSPHATE ION | | Authors: | Han, H, Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PXQ
 
 | | CDK2 in complex with 3 molecules of 8-anilino-1-naphthalene sulfonate | | Descriptor: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3EHJ
 
 | |
3EHH
 
 | |
3EHF
 
 | |
1HFS
 
 | |
3GIG
 
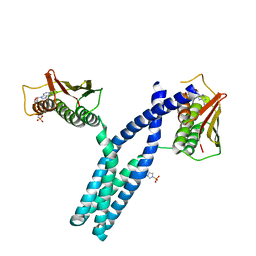 | | Crystal structure of phosphorylated DesKC in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GIF
 
 | | Crystal structure of DesKC_H188E in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GIE
 
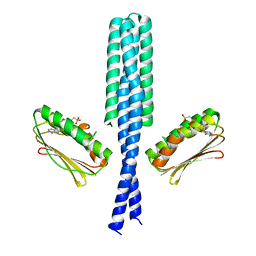 | | Crystal structure of DesKC_H188E in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EHG
 
 | |
4EZ3
 
 | | CDK2 in complex with NSC 134199 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(6-hydroxy-2-oxo-1,2-dihydropyridin-3-yl)diazenyl]benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Martin, M, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
7TGE
 
 | |
7TOU
 
 | |
7TOV
 
 | |
7TOX
 
 | |
7TOY
 
 | |
7TOZ
 
 | |
7TP0
 
 | |
