8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
7YP3
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with elaiophylin | | Descriptor: | ACETATE ION, Elaiophylin, GLYCEROL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4JN4
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7YP6
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with UDP | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP5
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with TDP | | Descriptor: | CHLORIDE ION, Glycosyltransferase, R-1,2-PROPANEDIOL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP4
 
 | | Crystal structure of elaiophylin glycosyltransferase in apo-form | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3I9O
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Graeff, R, Liu, Q, Kriksunov, I.A, Kotaka, M, Oppenheimer, N, Hao, Q, Lee, H.C. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38
J.Biol.Chem., 284, 2009
|
|
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JNF
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | Hsp70 CHAPERONE DnaK | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5K0X
 
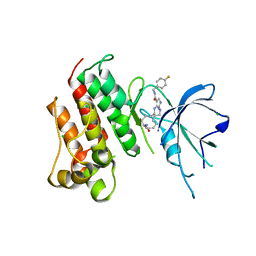 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
4WD7
 
 | |
3QB5
 
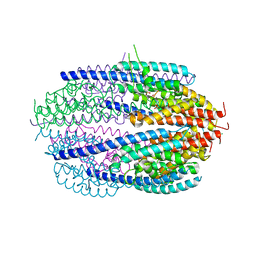 | | Human C3PO complex in the presence of MnSO4 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Ye, X, Huang, N, Liu, Y, Paroo, Z, Chen, S, Zhang, H, Liu, Q. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4ED5
 
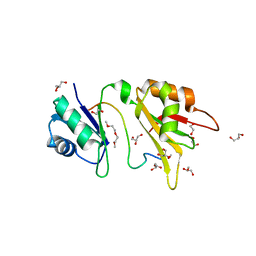 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4RAU
 
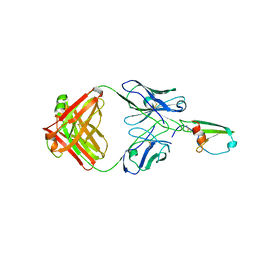 | |
4RYQ
 
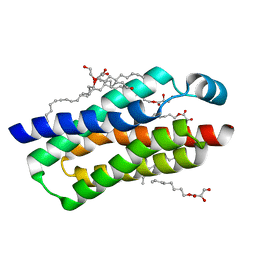 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom | | Descriptor: | Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYM
 
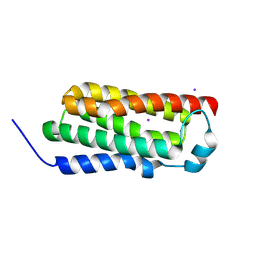 | |
4RYJ
 
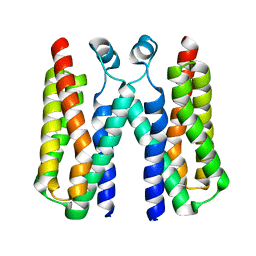 | |
4RYO
 
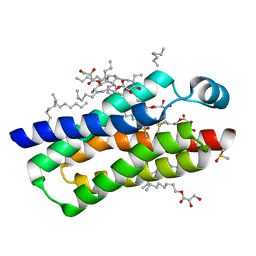 | | Crystal structure of BcTSPO type II high resolution monomer | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYR
 
 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom with DMSO | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RMK
 
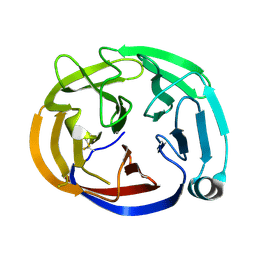 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | Descriptor: | CALCIUM ION, Latrophilin-3 | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4RYI
 
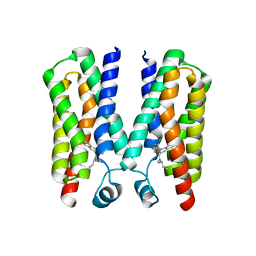 | |
4RYN
 
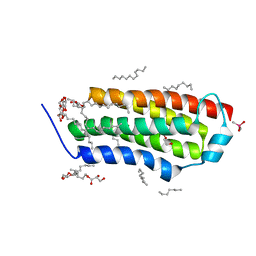 | | Crystal structure of BcTSPO, type1 monomer | | Descriptor: | CACODYLATE ION, DODECYL-ALPHA-D-MALTOSIDE, Integral membrane protein, ... | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RML
 
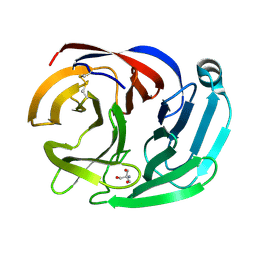 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
2INF
 
 | | Crystal Structure of Uroporphyrinogen Decarboxylase from Bacillus subtilis | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Fan, J, Liu, Q, Hao, Q, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis
J.Bacteriol., 189, 2007
|
|
4WD8
 
 | |
