6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LMS
 
 | |
7PYV
 
 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
6O3T
 
 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
5D1M
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (P199A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
5D1L
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (Y165A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, OXALATE ION, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
5D1K
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
4QUC
 
 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
6A0Z
 
 | | Crystal structure of broadly neutralizing antibody 13D4 bound to H5N1 influenza hemagglutinin, HA head region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 13D4, Fab Heavy Chain, ... | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
6A0X
 
 | | Crystal structure of broadly neutralizing antibody 13D4 | | Descriptor: | Antibody 13D4, Fab Heavy Chain, Fab Light Chain | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
3U28
 
 | | Crystal structure of a Cbf5-Nop10-Gar1 complex from Saccharomyces cerevisiae | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 3, H/ACA ribonucleoprotein complex subunit 4 | | Authors: | Ye, K, Li, S. | | Deposit date: | 2011-10-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase
Genes Dev., 25, 2011
|
|
3B2U
 
 | | Crystal structure of isolated domain III of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Ferguson, K.M, Li, S, Kussie, P. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.
Structure, 16, 2008
|
|
3B2V
 
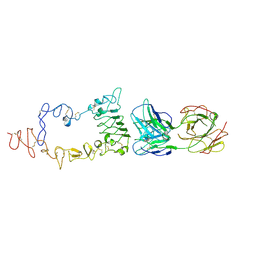 | | Crystal structure of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Ferguson, K.M, Li, S, Kussie, P. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.
Structure, 16, 2008
|
|
8JON
 
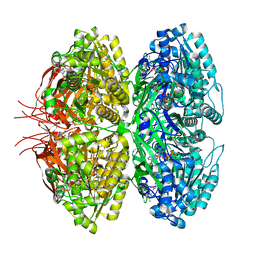 | | Structure of a synthetic circadian clock protein KaiC mutant of cyanobacteria Synechococcus elongatus PCC 7942 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, ... | | Authors: | Jia, X, Zhang, Q, Li, S, Guo, J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Adaptation of ancient cyanobacterial clock to the day length ~ 0.95 Ga ago
To Be Published
|
|
8HVS
 
 | |
8JEU
 
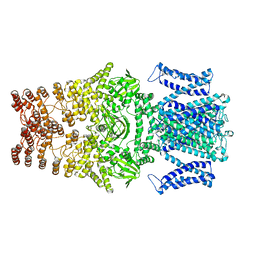 | |
8JEC
 
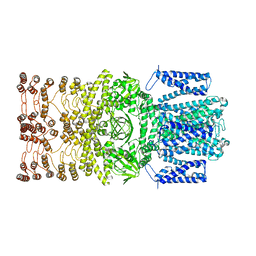 | |
8JET
 
 | |
5HYZ
 
 | | Crystal Structure of SCL7 in Oryza sativa | | Descriptor: | GRAS family transcription factor containing protein, expressed | | Authors: | Wu, Y, Li, S, Zhao, Y, Sun, L. | | Deposit date: | 2016-02-02 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa.
Plant Cell, 28, 2016
|
|
5GVU
 
 | |
7UXU
 
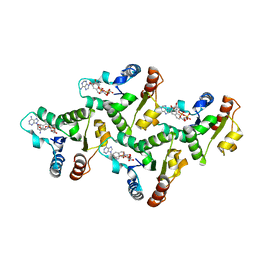 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | Descriptor: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
6JIP
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIQ
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
