1I9S
 
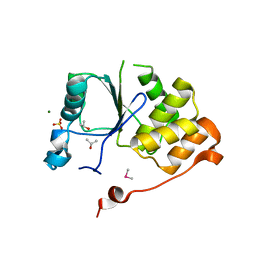 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
2O19
 
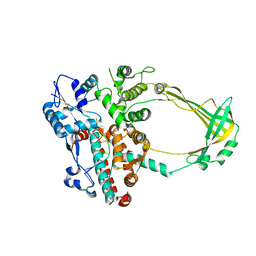 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R, Mondragon, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5E
 
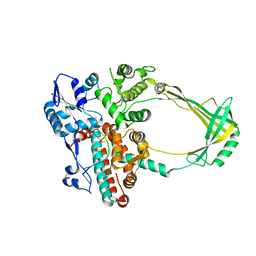 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O54
 
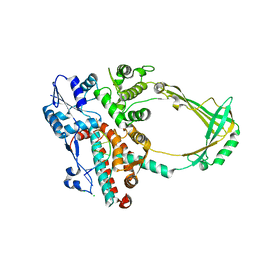 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, Digate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5C
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
1I7D
 
 | | NONCOVALENT COMPLEX OF E.COLI DNA TOPOISOMERASE III WITH AN 8-BASE SINGLE-STRANDED DNA OLIGONUCLEOTIDE | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA TOPOISOMERASE III, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule.
Nature, 411, 2001
|
|
2XEU
 
 | | Ring domain | | Descriptor: | RING FINGER PROTEIN 4, SULFATE ION, ZINC ION, ... | | Authors: | Plechanovova, A, McMahon, S.A, Johnson, K.A, Navratilova, I, Naismith, J.H, Hay, R.T. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of Ubiquitylation by Dimeric Ring Ligase Rnf4
Nat.Struct.Mol.Biol., 18, 2011
|
|
2O59
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol pH 8.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2N0R
 
 | |
1IAZ
 
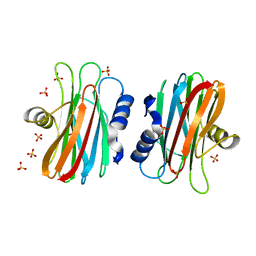 | | EQUINATOXIN II | | Descriptor: | EQUINATOXIN II, SULFATE ION | | Authors: | Athanasiadis, A, Anderluh, G, Macek, P, Turk, D. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the soluble form of equinatoxin II, a pore-forming toxin from the sea anemone Actinia equina.
Structure, 9, 2001
|
|
4JMQ
 
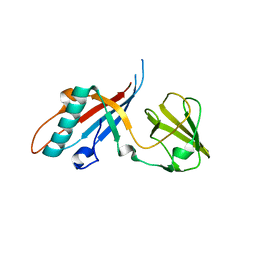 | | Crystal structure of pb9: The Dit of bacteriophage T5. | | Descriptor: | Bacteriophage T5 distal tail protein | | Authors: | Flayhan, A, Vellieux, F.M.D, Girard, E, Maury, O, Boulanger, P, Breyton, C. | | Deposit date: | 2013-03-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: a Conserved Structural Motif among All Siphophages.
J.Virol., 88, 2014
|
|
4KA4
 
 | |
7LSQ
 
 | |
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
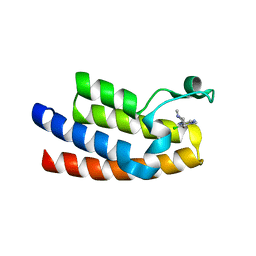 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRO
 
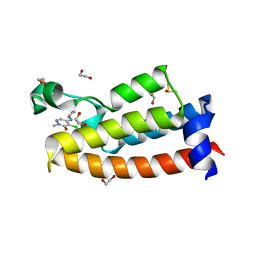 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7U15
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type B case 2 (case 7). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U16
 
 | | TMEM106B(120-254) protofilament from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (all cases combined). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U17
 
 | | TMEM106B(120-254) T185S singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type B case 2 (case 7). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U10
 
 | | TMEM106B(120-254) protofilament from progressive supranuclear palsy (PSP) case 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U11
 
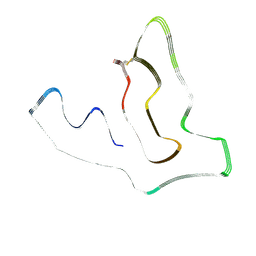 | | TMEM106B(120-254) protofilament from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U13
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
