6UZW
 
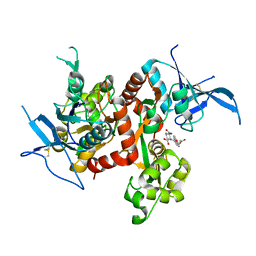 | | Crystal structure of GLUN1/GLUN2A ligand-binding domain in complex with glycine and UBP791 | | Descriptor: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Wang, J.X, Furukawa, H. | | Deposit date: | 2019-11-15 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
6VAK
 
 | | Cryo-EM structure of human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Syrjanen, J.L, Chou, T.H, Furukawa, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure and assembly of calcium homeostasis modulator proteins.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WHS
 
 | | GluN1b-GluN2B NMDA receptor in non-active 1 conformation at 3.95 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHV
 
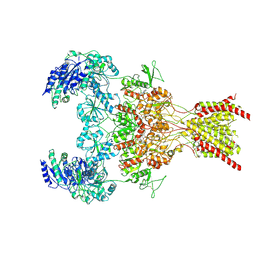 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHT
 
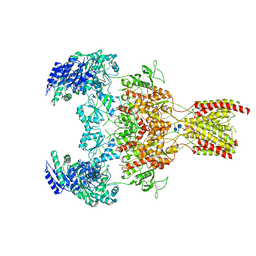 | | GluN1b-GluN2B NMDA receptor in active conformation at 4.4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WI0
 
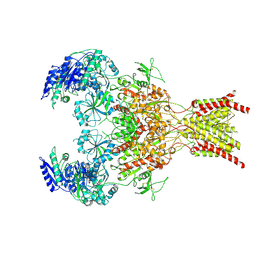 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WI1
 
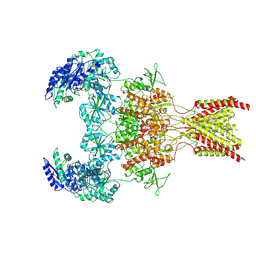 | | GluN1b-GluN2B NMDA receptor in active conformation stabilized by inter-GluN1b-GluN2B subunit cross-linking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHR
 
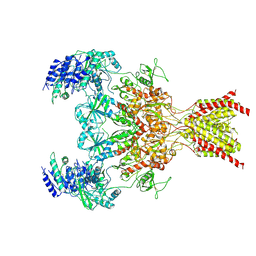 | | GluN1b-GluN2B NMDA receptor in non-active 2 conformation at 4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHX
 
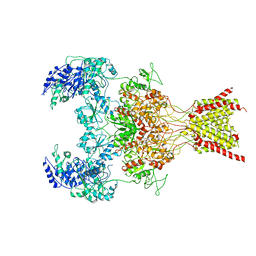 | | GluN1b-GluN2B NMDA receptor in complex with GluN2B antagonist SDZ 220-040, class 2 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHW
 
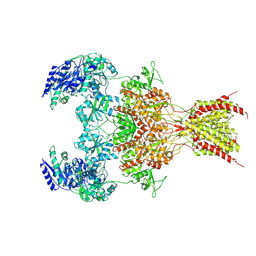 | | GluN1b-GluN2B NMDA receptor in complex with GluN2B antagonist SDZ 220-040, class 1 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHU
 
 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHY
 
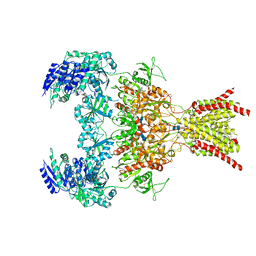 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
5FXJ
 
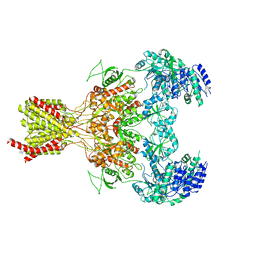 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5OJ6
 
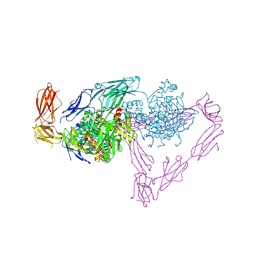 | | Crystal structure of the chicken MDGA1 ectodomain in complex with the human neuroligin 1 (NL1(-A-B)) cholinesterase domain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-1, ... | | Authors: | Elegheert, J, Clayton, A.J, Aricescu, A.R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Mechanism for Modulation of Synaptic Neuroligin-Neurexin Signaling by MDGA Proteins.
Neuron, 95, 2017
|
|
5OJ2
 
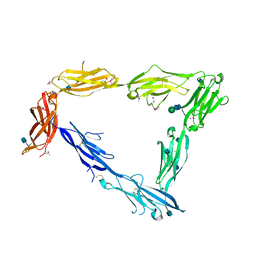 | | Crystal structure of the chicken MDGA1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clayton, A.J, Aricescu, A.R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Mechanism for Modulation of Synaptic Neuroligin-Neurexin Signaling by MDGA Proteins.
Neuron, 95, 2017
|
|
5OJK
 
 | |
4R0W
 
 | |
1Y20
 
 | |
1Y1M
 
 | |
1Y1Z
 
 | |
4KFQ
 
 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | Descriptor: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
4R0U
 
 | |
4R0P
 
 | |
8IE2
 
 | | Crystal structure of Lactiplantibacillus plantarum GlyRS | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2023-02-15 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of tRNA recognition by heterotetrameric glycyl-tRNA synthetase from lactic acid bacteria.
J.Biochem., 174, 2023
|
|
