7Q4I
 
 | | Crystal structure of DmC1GalT1 in complex with UDP-Mn2+ and the APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1, ... | | Authors: | Gonzalez-Ramirez, A.M, Coelho, H, Companon, I, Grosso, A.S, Yang, Z, Narimatsu, Y, Clausen, H, Marcelo, F, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2021-10-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the synthesis of the core 1 structure by C1GalT1.
Nat Commun, 13, 2022
|
|
7A06
 
 | | Structure of human CKa1 in complex with compound o | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-phenylphenyl)methyl]-4-pyrrolidin-1-yl-pyridine, Choline kinase alpha | | Authors: | Serran-Aguilera, L, Mariotto, E, Rubbini, G, Castro Navas, F.C, Marco, C, Carrasco-Jimenez, M.P, Ballarotto, M, Macchiarulo, A, Hurtado-Guerrero, R, Viola, G, Lopez-Cara, L.C. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, biological evaluation, in silico modeling and crystallization of novel small monocationic molecules with potent antiproliferative activity by dual mechanism.
Eur.J.Med.Chem., 207, 2020
|
|
5FIH
 
 | | SACCHAROMYCES CEREVISIAE GAS2P (E176Q MUTANT) IN COMPLEX WITH LAMINARITETRAOSE AND LAMINARIPENTAOSE | | Descriptor: | 1,3-BETA-GLUCANOSYLTRANSFERASE, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Raich, L, Borodkin, V, van Aalten, D.M.F, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Trapped Covalent Intermediate of a Glycoside Hydrolase on the Pathway to Transglycosylation. Insights from Experiments and Quantum Mechanics/Molecular Mechanics Simulations.
J. Am. Chem. Soc., 138, 2016
|
|
5FUT
 
 | | Human choline kinase a1 in complex with compound 4-(dimethylamino)-1-{4-[4-(4-{[4-(pyrrolidin- 1-yl)pyridinium-1-yl]methyl}phenyl)butyl]benzyl}pyridinium (compound BR25) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-1-{4-[4-(4-{[4-(pyrrolidin-1-yl)pyridinium-1-yl]methyl}phenyl)butyl]benzyl}pyridinium, CHOLINE KINASE ALPHA | | Authors: | Serran-Aguilera, L, Denton, H, Rubio-Ruiz, B, Lopez-Gutierrez, B, Entrena, A, Izquierdo, L, Smith, T.K, Conejo-Garcia, A, Hurtado-Guerrero, R. | | Deposit date: | 2016-01-29 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasmodium Falciparum Choline Kinase Inhibition Leads to a Major Decrease in Phosphatidylethanolamine Causing Parasite Death.
Sci.Rep., 6, 2016
|
|
6TGG
 
 | | scFv-1SM3 in complex with glycopeptide containing an sp2-imino sugar | | Descriptor: | 1,2-ETHANEDIOL, Mucin-1, ScFv_SM3, ... | | Authors: | Bermejo, I.A, Navo, C.D, Castro-Lopez, J, Samchez-Fernandez, E.M, Guerreiro, A, Avenoza, A, Busto, J.H, Garcia-Martin, F, Nishimura, S.I, Garcia-Fernandez, J.M, Ortiz-Mellet, C, Bernardes, G.J.L, Hurtado-Guerrero, R, Peregrina, J.M, Corzana, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp 2 -iminosugar fragment.
Chem Sci, 11, 2020
|
|
4R3R
 
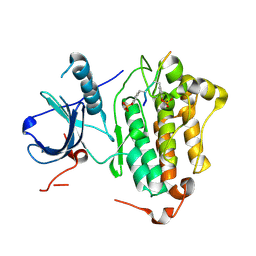 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7KVC
 
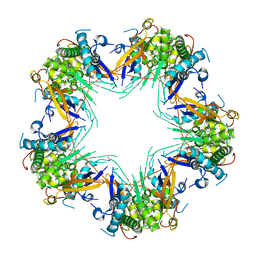 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
6YUG
 
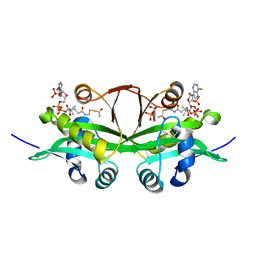 | | Crystal structure of C. parvum GNA1 in complex with acetyl-CoA and glucose 6P. | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ACETYL COENZYME *A, Diamine acetyltransferase | | Authors: | Chi, J, Cova, M, de las Rivas, M, Medina, A, Borges, R, Leivar, P, Planas, A, Uson, I, Hurtado-Guerrero, R, Izquierdo, L. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plasmodium falciparum Apicomplexan-Specific Glucosamine-6-Phosphate N -Acetyltransferase Is Key for Amino Sugar Metabolism and Asexual Blood Stage Development.
Mbio, 11, 2020
|
|
4R3P
 
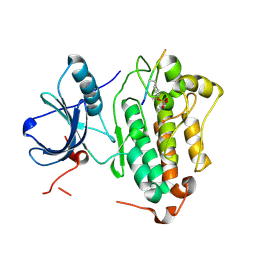 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3ZY5
 
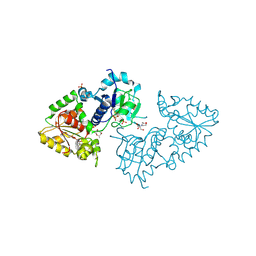 | | Crystal structure of POFUT1 in complex with GDP-fucose (crystal-form-I) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1, ... | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
3ZY3
 
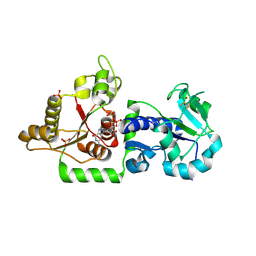 | | Crystal structure of POFUT1 in complex with GDP (crystal-form-III) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
3ZY2
 
 | | Crystal structure of POFUT1 in complex with GDP (High resolution dataset) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1 | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
3ZY4
 
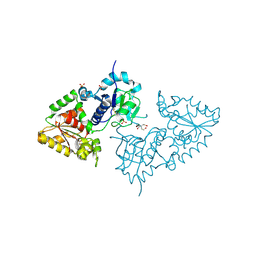 | | Crystal structure of POFUT1 apo-form (crystal-form-I) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
3ZY6
 
 | | Crystal structure of POFUT1 in complex with GDP-fucose (crystal-form-II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1 | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
6F0F
 
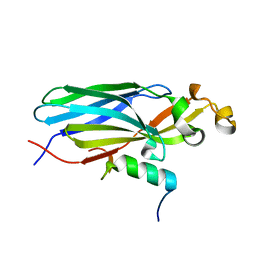 | | Crystal structure ASF1-ip2_s | | Descriptor: | Histone chaperone ASF1A, ip2_s | | Authors: | Gaubert, A, Guichard, B, Murciano, B, Le Du, M.H, Ochsenbein, F, Guerois, R, Andreani, J. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6F0G
 
 | | Crystal structure ASF1-ip3 | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, ip3 | | Authors: | Gaubert, A, Guichard, B, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6F0H
 
 | | Crystal structure ASF1-ip4 | | Descriptor: | CITRIC ACID, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6G9V
 
 | | Crystal structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase(AfUAP1) in complex with UDPGlcNAc, pyrophosphate and Mg2+ | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, UDP-N-acetylglucosamine pyrophosphorylase, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
6G9W
 
 | | Crystal Structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase in complex with UTP | | Descriptor: | UDP-N-acetylglucosamine pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
6YIG
 
 | | Crystal structure of the N-terminal EF-hand domain of Arabidopsis thaliana AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein, SODIUM ION | | Authors: | Yperman, K, Merceron, R, De Munck, S, Bloch, Y, Savvides, S.N, Pleskot, R, Van Damme, D. | | Deposit date: | 2020-04-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
8AXH
 
 | | Crystal structure of a MUC1-like glycopeptide containing the unnatural L-4-hydroxynorvaline in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Mucin-1 subunit alpha, ... | | Authors: | Bermejo, I, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2022-08-31 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Approach for the Development of MUC1-Glycopeptide-Based Cancer Vaccines with Predictable Responses.
Jacs Au, 4, 2024
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
