6GBK
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
6GBL
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
6GBJ
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
6FHF
 
 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Design, SODIUM ION | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
6GC2
 
 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
8OV0
 
 | | MUC5AC CysD7 amino acids 3518-3626 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mucin-5AC | | Authors: | Khmelnitsky, L, Milo, A, Dym, O, Fass, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diversity of CysD domains in gel-forming mucins.
Febs J., 290, 2023
|
|
6FPR
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6FPK
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6HAP
 
 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
1D3A
 
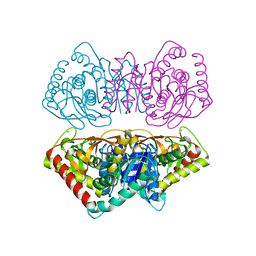 | | CRYSTAL STRUCTURE OF THE WILD TYPE HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, HALOPHILIC MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-09-28 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
1S0W
 
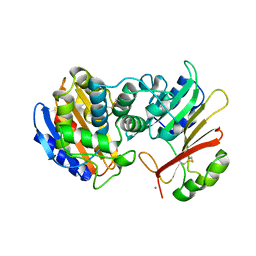 | |
2HLP
 
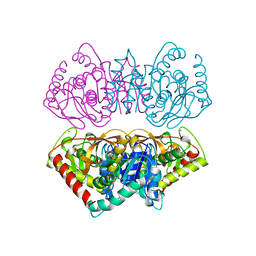 | | CRYSTAL STRUCTURE OF THE E267R MUTANT OF A HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
8QCI
 
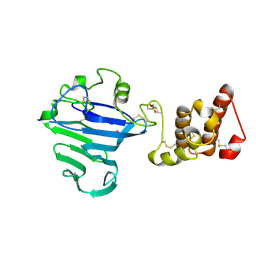 | | FCGBP D10 Assembly Segment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Yeshaya, N, Fass, D. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | VWD domain stabilization by autocatalytic Asp-Pro cleavage.
Protein Sci., 33, 2024
|
|
3FPL
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Felix, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
3FPC
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-294 of T. brockii ADH by E. histolytica ADH | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, IMIDAZOLE, ... | | Authors: | Felix, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
3FSR
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NADP-dependent alcohol dehydrogenase, ... | | Authors: | Frolow, F, Greenblatt, H, Goihberg, E, Burstein, Y. | | Deposit date: | 2009-01-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
3FTN
 
 | | Q165E/S254K Double Mutant Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms.
Biochemistry, 49, 2010
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
4P3D
 
 | | MT1-MMP:Fab complex (Form II) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Rozenberg, H, Udi, Y, Sagi, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
4P3C
 
 | | MT1-MMP:Fab complex (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Rozenberg, H, Udi, Y, Sagi, I. | | Deposit date: | 2014-03-06 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
1O6Z
 
 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
4PTN
 
 | | Crystal Structure of YagE, a KDG aldolase protein in complex with Magnesium cation coordinated L-glyceraldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-glyceraldehyde, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12.
Proteins, 71, 2008
|
|
3QA9
 
 | |
4JCO
 
 | |
3TPJ
 
 | | APO structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
