6S1V
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7LMX
 
 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7MEZ
 
 | | Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 5 | | Authors: | Burke, J.E, Dalwadi, U, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the phosphoinositide 3-kinase (PI3K) p110 gamma-p101 complex reveals molecular mechanism of GPCR activation.
Sci Adv, 7, 2021
|
|
5IZS
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 5L6HC3_1 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Oberdorfer, G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6BAA
 
 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Martin, G.M, Yoshioka, C, Shyng, S.L. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-01 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
8TCF
 
 | | Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8TCG
 
 | | Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Integrin alpha-V heavy chain, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8SK7
 
 | | Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA_20 minibinder (RFdiffusion-designed), ... | | Authors: | Borst, A.J, Bennett, N.R. | | Deposit date: | 2023-04-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | De novo design of protein structure and function with RFdiffusion.
Nature, 620, 2023
|
|
8OOK
 
 | |
8OOR
 
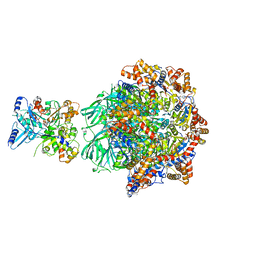 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
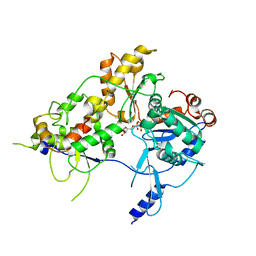 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | Descriptor: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOC
 
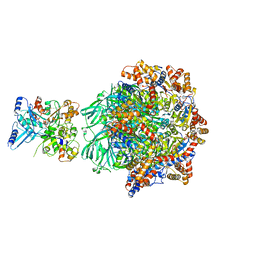 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MSP
 
 | | De novo Designed Protein Foldit3 | | Descriptor: | De novo Designed Protein Foldit3 | | Authors: | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | Deposit date: | 2018-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MRS
 
 | |
3Q6O
 
 | | Oxidoreductase Fragment of Human QSOX1 | | Descriptor: | SULFATE ION, Sulfhydryl oxidase 1 | | Authors: | Fass, D, Alon, A. | | Deposit date: | 2011-01-03 | | Release date: | 2012-05-30 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
3QD9
 
 | |
