8GS1
 
 | | Crystal structure of AziU2-U3 complex from Streptomyces sahachiroi NRRL2485 | | Descriptor: | Azi28, Azi29, FORMIC ACID, ... | | Authors: | Cheng, Y, Li, P, Liu, W, Fang, P. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
8HK0
 
 | | Crystal structure of Fic32-33 complex from Streptomyces ficellus NRRL 8067 | | Descriptor: | Acyl-CoA dehydrogenase, Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cheng, Y, Qiao, H, Liu, W, Fang, P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
5NPK
 
 | | 1.98A STRUCTURE OF THIOPHENE1 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit B,DNA gyrase subunit B,DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-17 | | Release date: | 2017-07-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8P5Y
 
 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Z
 
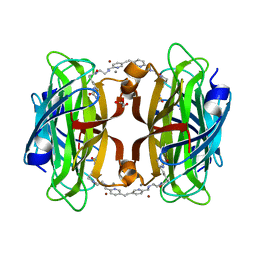 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4R3M
 
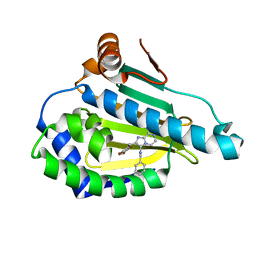 | | Crystal structure of Human Hsp90 with JR9 | | Descriptor: | Heat shock protein HSP 90-alpha, N~3~-benzyl-2-[(6-bromo-1,3-benzodioxol-5-yl)methyl]imidazo[1,2-a]pyrazine-3,8-diamine | | Authors: | Li, J, Yang, M, Ren, J, Xiong, B, He, J. | | Deposit date: | 2014-08-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multi-substituted 8-aminoimidazo[1,2-a]pyrazines by Groebke-Blackburn-Bienayme reaction and their Hsp90 inhibitory activity.
Org.Biomol.Chem., 13, 2015
|
|
6O5I
 
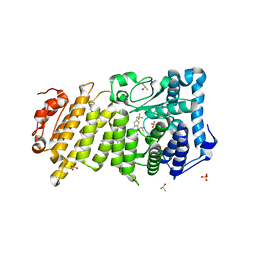 | | Menin in complex with MI-3454 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, SULFATE ION, ... | | Authors: | Linhares, B.M, Klossowski, S, Cierpicki, T, Grembecka, J. | | Deposit date: | 2019-03-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24025619 Å) | | Cite: | Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia.
J.Clin.Invest., 130, 2020
|
|
6OCO
 
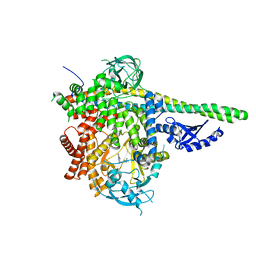 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 4-[(1S,4S)-5-(3-chlorophenyl)-2,5-diazabicyclo[2.2.1]heptan-2-yl]-2-(pyridin-3-yl)pyrimidine-5-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery and optimization of heteroaryl piperazines as potent and selective PI3K delta inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4L8Z
 
 | | Crystal structure of Human Hsp90 with RL1 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](3,4-dihydroisoquinolin-2(1H)-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3B8R
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a naphthamide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-cyclopropyl-6-[(6,7-dimethoxyquinolin-4-yl)oxy]naphthalene-1-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Gu, Y, Zhao, H. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evaluation of a Series of Naphthamides as Potent, Orally Active Vascular
Endothelial Growth Factor Receptor-2 Tyrosine Kinase Inhibitors
J.Med.Chem., 51, 2008
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | Descriptor: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
5KZ8
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 5,5-dimethyl-7-[(1~{S})-4-oxidanyl-1~{H}-inden-1-yl]-2-phenylazanyl-pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5KZ7
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 7-[(1~{S})-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(pyridin-3-ylamino)pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4L08
 
 | | Crystal structure of the maleamate amidase Ami(C149A) in complex with maleate from Pseudomonas putida S16 | | Descriptor: | Hydrolase, isochorismatase family, MALEIC ACID | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4L07
 
 | | Crystal structure of the maleamate amidase Ami from Pseudomonas putida S16 | | Descriptor: | GLYCEROL, Hydrolase, isochorismatase family, ... | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4P59
 
 | | HER3 extracellular domain in complex with Fab fragment of MOR09825 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOR09825 Fab fragment heavy chain, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin.
Cancer Res., 73, 2013
|
|
3B8Q
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a naphthamide inhibitor | | Descriptor: | N-(4-chlorophenyl)-6-[(6,7-dimethoxyquinolin-4-yl)oxy]naphthalene-1-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Gu, Y, Zhao, H. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Naphthamides as Novel and Potent Vascular Endothelial Growth Factor
Receptor Tyrosine Kinase Inhibitors: Design, Synthesis and Evaluation
J.Med.Chem., 51, 2008
|
|
3BE2
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzamide inhibitor | | Descriptor: | N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Gu, Y, Rose, P, Zhao, H. | | Deposit date: | 2007-11-16 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Naphthamides as novel and potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: design, synthesis, and evaluation.
J.Med.Chem., 51, 2008
|
|
3DOR
 
 | | Crystal Structure of mature CPAF | | Descriptor: | Protein CT_858, SULFATE ION | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-06 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
 | | Structure of mature CPAF complexed with lactacystin | | Descriptor: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DJA
 
 | | Crystal Structure of cpaf solved with MAD | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-06-22 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPN
 
 | | Crystal Structure of cpaf s499a mutant | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
5EAK
 
 | |
8ETN
 
 | |
