3T0W
 
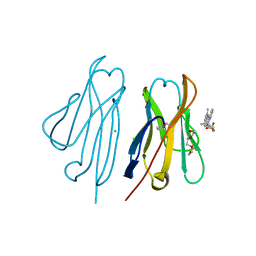 | | Fluorogen activating protein M8VL in complex with dimethylindole red | | Descriptor: | 1-(3-sulfopropyl)-4-[(1E,3E)-3-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)prop-1-en-1-yl]quinolinium, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
3T0X
 
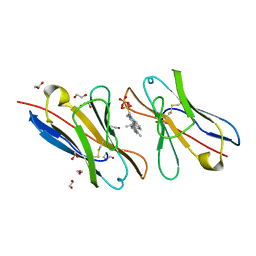 | | Fluorogen Activating Protein M8VLA4(S55P) in complex with dimethylindole red | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-sulfopropyl)-4-[(1E,3E)-3-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)prop-1-en-1-yl]quinolinium, Immunoglobulin variable lambda domain M8VLA4(S55P), ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
3T0V
 
 | | Unliganded fluorogen activating protein M8VL | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
2KGN
 
 | |
3T0J
 
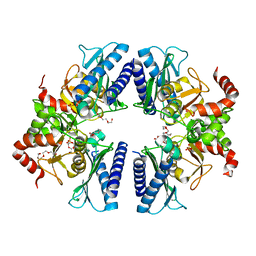 | |
2HXW
 
 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
2KNS
 
 | |
2L36
 
 | |
2LKE
 
 | | Structures and Interaction Analyses of the Integrin Alpha-M Beta-2 Cytoplasmic Tails | | Descriptor: | Integrin alpha-M | | Authors: | Chua, G.L, Tang, X.Y, Amalraj, M, Bhattacharjya, S, Tan, S.M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures and interaction analyses of the integrin alphaMbeta2 cytoplasmic tails
J.Biol.Chem., 2011
|
|
2LM8
 
 | |
2LKJ
 
 | | Structures and Interaction Analyses of the Integrin Alpha-M Beta-2 Cytoplasmic Tails | | Descriptor: | Integrin alpha-M | | Authors: | Chua, G.L, Tang, X.Y, Amalraj, M, Tan, S.M, Bhattacharjya, S. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-02 | | Last modified: | 2011-11-16 | | Method: | SOLUTION NMR | | Cite: | Structures and interaction analyses of the integrin alphaMbeta2 cytoplasmic tails
J.Biol.Chem., 2011
|
|
2L4U
 
 | |
2LUV
 
 | |
2MQ2
 
 | |
2MQ4
 
 | |
2MQ5
 
 | |
7EVR
 
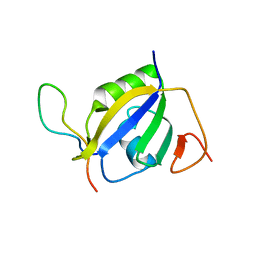 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVS
 
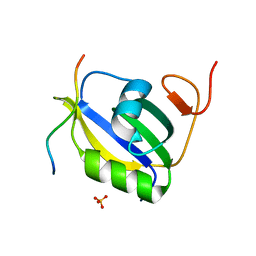 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
6TWA
 
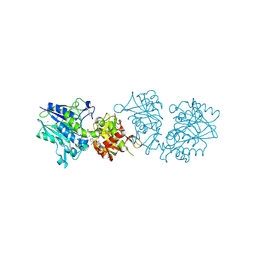 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12646 (an AOPCP derivative, compound 20 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-19 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TW0
 
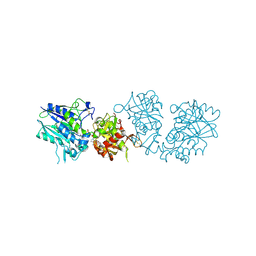 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12690 (an AOPCP derivative, compound 10 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-2-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TWF
 
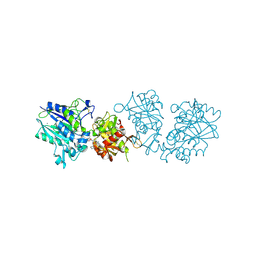 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12604 (an AOPCP derivative, compound 21 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TVX
 
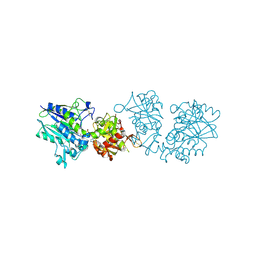 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12676 (an AOPCP derivative, compound 9 in paper) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TVE
 
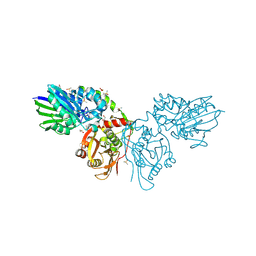 | | Unliganded human CD73 (5'-nucleotidase) in the open state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Scaletti, E, Strater, N. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TVG
 
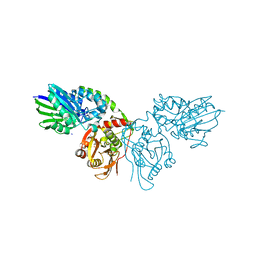 | |
6S7H
 
 | | Human CD73 (5'-nucleotidase) in complex with PSB12489 (an AOPCP derivative) in the closed state | | Descriptor: | (N6,N6)-methyl,benzyl-C2-chloro-(alpha,beta)-methylene-ADP, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-Ray Co-Crystal Structure Guides the Way to Subnanomolar Competitive Ecto-5'-Nucleotidase (CD73) Inhibitors for Cancer Immunotherapy
Adv Ther, 2019
|
|
