2LHL
 
 | | Chemical Shift Assignments and solution structure of human apo-S100A1 E32Q mutant | | Descriptor: | Protein S100-A1 | | Authors: | Ruszczynska-Bartnik, K, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N NMR sequence-specific resonance assignments and relaxation parameters for human apo-S100A1 E32Q mutant
To be Published
|
|
2M3W
 
 | |
5OH6
 
 | | Legionella pneumophila RidL N-terminal domain lacking beta hairpin | | Descriptor: | Interaptin | | Authors: | Baerlocher, K, Hutter, C.A.J, Swart, A.L, Steiner, B, Welin, A, Hohl, M, Letourneur, F, Seeger, M.A, Hilbi, H. | | Deposit date: | 2017-07-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into Legionella RidL-Vps29 retromer subunit interaction reveal displacement of the regulator TBC1D5.
Nat Commun, 8, 2017
|
|
5OH5
 
 | | Legionella pneumophila RidL N-terminal retromer binding domain | | Descriptor: | RidL | | Authors: | Baerlocher, K, Hutter, C.A.J, Swart, A.L, Steiner, B, Welin, A, Hohl, M, Letourneur, F, Seeger, M.A, Hilbi, H. | | Deposit date: | 2017-07-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into Legionella RidL-Vps29 retromer subunit interaction reveal displacement of the regulator TBC1D5.
Nat Commun, 8, 2017
|
|
2RDF
 
 | |
4ACP
 
 | | Deactivation of human IgG1 Fc by endoglycosidase treatment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG GAMMA-1 CHAIN C REGION | | Authors: | Raman, K, Bowden, T.A, Krishna, B.A, Dwek, R.A, Crispin, M, Scanlan, C.N. | | Deposit date: | 2011-12-16 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Selective Deactivation of Serum Igg: A General Strategy for the Enhancement of Monoclonal Antibody Receptor Interactions.
J.Mol.Biol., 420, 2012
|
|
3LTQ
 
 | |
1A3P
 
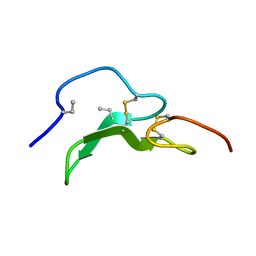 | | ROLE OF THE 6-20 DISULFIDE BRIDGE IN THE STRUCTURE AND ACTIVITY OF EPIDERMAL GROWTH FACTOR, NMR, 20 STRUCTURES | | Descriptor: | EPIDERMAL GROWTH FACTOR | | Authors: | Barnham, K, Torres, A, Alewood, D, Alewood, P, Domagala, T, Nice, E, Norton, R. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-29 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Role of the 6-20 disulfide bridge in the structure and activity of epidermal growth factor.
Protein Sci., 7, 1998
|
|
6R0V
 
 | |
6R0U
 
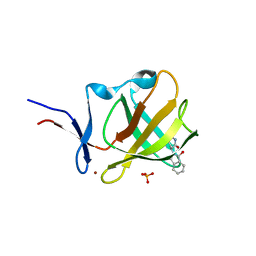 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5a and hydrolysis product | | Descriptor: | 3-azanyl-2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]benzoic acid, 4-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0S
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 4a and hydrolysis product | | Descriptor: | 2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]-4-nitro-isoindole-1,3-dione, 2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]-6-nitro-benzoic acid, CEREBLON ISOFORM 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0Q
 
 | |
6R11
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5b | | Descriptor: | 5-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, Cereblon isoform 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R13
 
 | |
6R12
 
 | |
6R18
 
 | |
6R19
 
 | |
6R1A
 
 | |
6R1D
 
 | |
6R1K
 
 | |
6R1W
 
 | |
6R1X
 
 | |
6R1C
 
 | |
6H9L
 
 | |
6H9M
 
 | | Coiled-coil domain-containing protein 90B residues 43-125 from Homo sapiens fused to a GCN4 adaptor | | Descriptor: | Coiled-coil domain-containing protein 90B, mitochondrial,General control protein GCN4 | | Authors: | Adlakha, J, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2018-08-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of MCU-Binding Proteins MCUR1 and CCDC90B - Representatives of a Protein Family Conserved in Prokaryotes and Eukaryotic Organelles.
Structure, 27, 2019
|
|
