7EL9
 
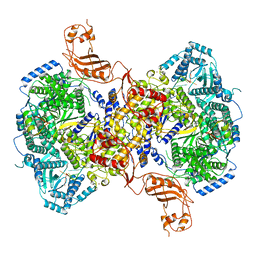 | | Structure of Machupo virus L polymerase in complex with Z protein and 3'-vRNA (dimeric complex) | | Descriptor: | MANGANESE (II) ION, Machupo virus 3'-vRNA promoter, RING finger protein Z, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
7ELB
 
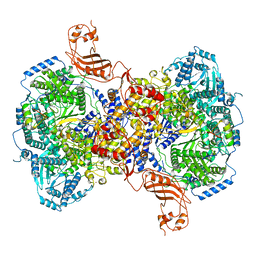 | | Structure of Machupo virus L polymerase in complex with Z protein (dimeric form) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
8K9E
 
 | |
8K9F
 
 | |
5WA5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD11-50 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2017-06-24 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
4HFO
 
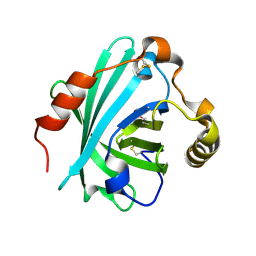 | | Biogenic amine-binding protein selenomethionine derivative | | Descriptor: | Biogenic amine-binding protein | | Authors: | Andersen, J.F, Xu, X, Chang, B, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GE1
 
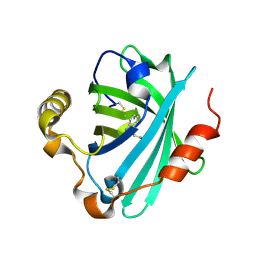 | | Structure of the tryptamine complex of the amine binding protein of Rhodnius prolixus | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Biogenic amine-binding protein, GLYCEROL | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GET
 
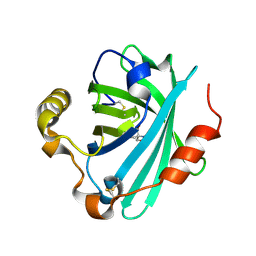 | | Crystal structure of biogenic amine binding protein from Rhodnius prolixus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Biogenic amine-binding protein | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8J5O
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 100lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8J5P
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 2,000lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8JKZ
 
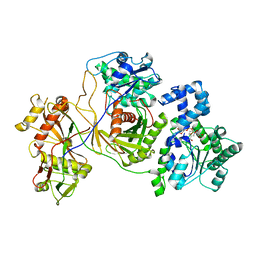 | |
8JL0
 
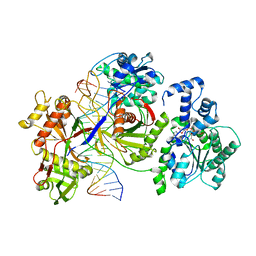 | | Cryo-EM structure of the prokaryotic SPARSA system complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain protein, ... | | Authors: | Xu, X, Zhen, X, Long, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antiphage immunity generated by a prokaryotic Argonaute-associated SPARSA system.
Nat Commun, 15, 2024
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
5SUJ
 
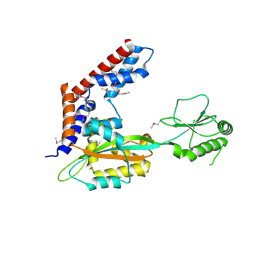 | | Crystal structure of uncharacterized protein LPG2148 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila.
Cell Rep, 23, 2018
|
|
7AL0
 
 | | Crystal Structure of Heymonin, a Novel Frog-derived Peptide | | Descriptor: | CHLORIDE ION, Heymonin | | Authors: | Kascakova, B, Prudnikova, T, Kuta Smatanova, I, Xu, X. | | Deposit date: | 2020-10-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization and functional analysis of cathelicidin-MH, a novel frog-derived peptide with anti-septicemic properties.
Elife, 10, 2021
|
|
2RHD
 
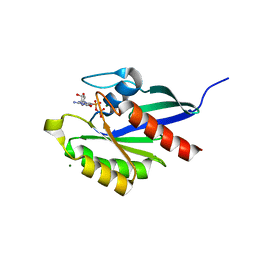 | | Crystal structure of Cryptosporidium parvum small GTPase RAB1A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Small GTP binding protein rab1a | | Authors: | Dong, A, Xu, X, Lew, J, Lin, Y.H, Khuu, C, Sun, X, Qiu, W, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of Cryptosporidium parvum small GTPase RAB1A.
To be Published
|
|
2R9Q
 
 | | Crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from Agrobacterium tumefaciens | | Descriptor: | 2'-deoxycytidine 5'-triphosphate deaminase, Synthetic peptide 1, Synthetic peptide 2 | | Authors: | Zhang, R, Dong, A, Xu, X, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from Agrobacterium tumefaciens.
To be Published
|
|
2RAQ
 
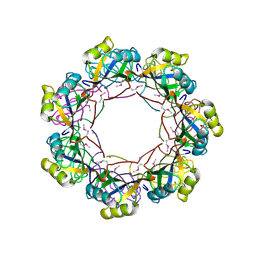 | | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus. Northeast Structural Genomics Consortium target TT205 | | Descriptor: | CALCIUM ION, Conserved protein MTH889 | | Authors: | Forouhar, F, Su, M, Xu, X, Seetharaman, J, Mao, L, Xiao, R, Ma, L.-C, Conover, K, Baran, M.C, Acton, T.B, Montelione, G.T, Arrowsmith, C.H, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-17 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus.
To be Published
|
|
2REK
 
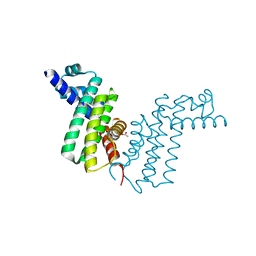 | | Crystal structure of tetR-family transcriptional regulator | | Descriptor: | ACETATE ION, Putative tetR-family transcriptional regulator | | Authors: | Dong, A, Xu, X, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of tetR-family transcriptional regulator.
To be Published
|
|
2RFL
 
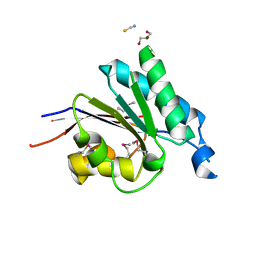 | | Crystal structure of the putative phosphohistidine phosphatase SixA from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase SixA, ... | | Authors: | Kim, Y, Binkowski, T, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Putative Phosphohistidine Phosphatase SixA from
Agrobacterium tumefaciens.
To be Published
|
|
3D01
 
 | | Crystal structure of the protein Atu1372 with unknown function from Agrobacterium tumefaciens | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Uncharacterized protein | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-30 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the protein Atu1372 with unknown function from Agrobacterium tumefaciens.
To be Published
|
|
3DED
 
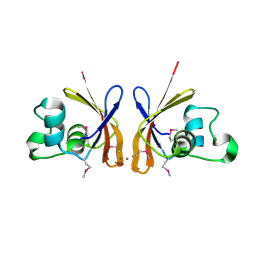 | | C-terminal domain of Probable hemolysin from Chromobacterium violaceum | | Descriptor: | CALCIUM ION, Probable hemolysin | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of C-terminal domain of Probable hemolysin from Chromobacterium violaceum
To be Published
|
|
3DCI
 
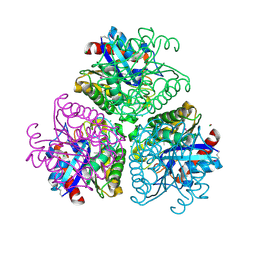 | | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Arylesterase, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Binkowski, T.A, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58
TO BE PUBLISHED
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
