8IQF
 
 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQG
 
 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7K6Q
 
 | | Active state Dot1 bound to the H4K16ac nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7K6P
 
 | | Active state Dot1 bound to the unacetylated H4 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7JZV
 
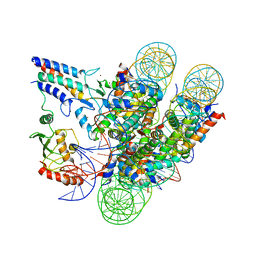 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5YWX
 
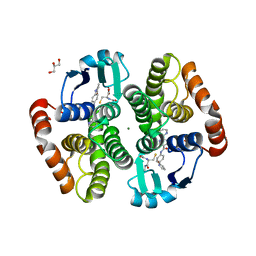 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with F092 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Omura, A, Tanaka, A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of crystal water molecules in a high-affinity inhibitor and hematopoietic prostaglandin D synthase complex by interaction energy studies.
Bioorg. Med. Chem., 26, 2018
|
|
5EUV
 
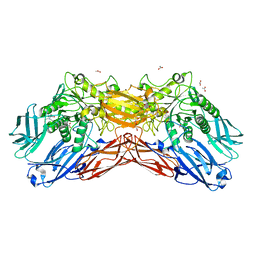 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2015-11-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6K1S
 
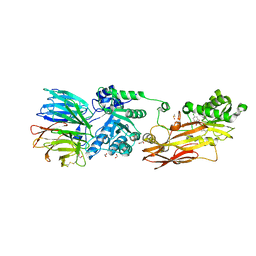 | | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase (PRMT5) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-[[7-[(2~{R},3~{R},4~{S},5~{R})-5-[(~{R})-(4-chlorophenyl)-oxidanyl-methyl]-3,4-bis(oxidanyl)oxolan-2-yl]pyrrolo[2,3-d]pyrimidin-4-yl]amino]ethanal, DIMETHYL SULFOXIDE, ... | | Authors: | Tong, S, Lin, H. | | Deposit date: | 2019-05-12 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7V4Y
 
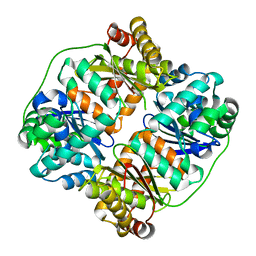 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YBH
 
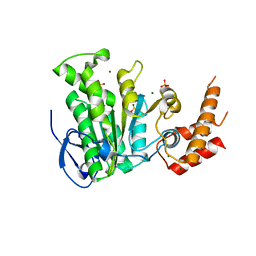 | |
5YKF
 
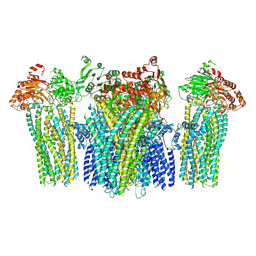 | |
5XZR
 
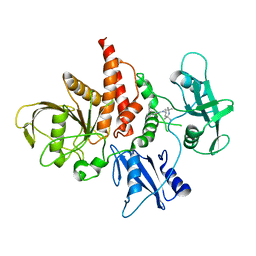 | | The atomic structure of SHP2 E76A mutant in complex with allosteric inhibitor 9b | | Descriptor: | 4-(3-phenylphenyl)-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3-thiazol-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Li, D, Xie, J, Zhu, J, Liu, C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Inhibitors of SHP2 with Therapeutic Potential for Cancer Treatment.
J. Med. Chem., 60, 2017
|
|
5YKG
 
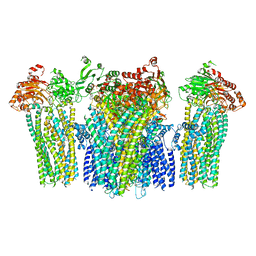 | |
5YDP
 
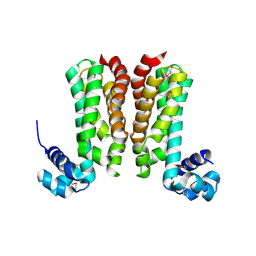 | |
5YBI
 
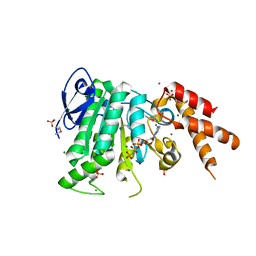 | | Structure of the bacterial pathogens ATPase with substrate AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Mu, Z.X, Gao, X.P, Cui, S. | | Deposit date: | 2017-09-05 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri.
Front Microbiol, 9, 2018
|
|
5YKE
 
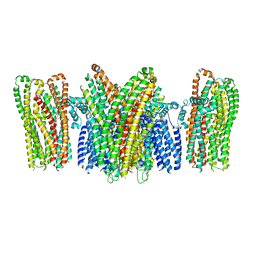 | |
5YTX
 
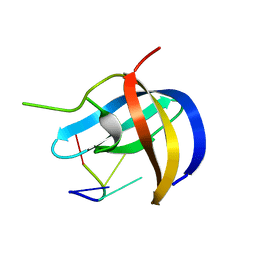 | | Crystal structure of YB1 cold-shock domain in complex with UCAACU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*AP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
4OGX
 
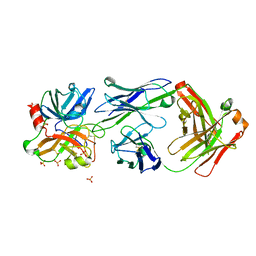 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.4 Angstrom resolution | | Descriptor: | DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, Plasma kallikrein, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
5YTT
 
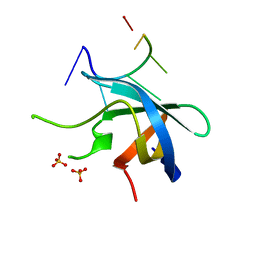 | | Crystal structure of YB1 cold-shock domain in complex with UCAUGU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*GP*U)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTS
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCUUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*CP*UP*UP*C)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTV
 
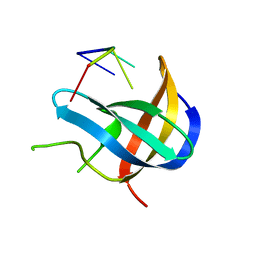 | | Crystal structure of YB1 cold-shock domain in complex with UCAUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YW9
 
 | |
5YWB
 
 | |
5YWC
 
 | |
