1WBS
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-5-MORPHOLIN-4-YL-N-[3-(2-PYRIDIN-4-YLETHYL)-1H-INDOL-5-YL]BENZAMIDE, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
1WBN
 
 | | fragment based p38 inhibitors | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-TERT-BUTYL-1H-PYRAZOL-5-YL)-N'-{4-CHLORO-3-[(PYRIDIN-3-YLOXY)METHYL]PHENYL}UREA | | Authors: | Cleasby, A, Devine, L.A, Gill, A.L, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
2R9E
 
 | |
1WBV
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-N-1H-INDOL-5-YL-5-MORPHOLIN-4-YLBENZAMIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
2TSA
 
 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1WWA
 
 | | NGF BINDING DOMAIN OF HUMAN TRKA RECEPTOR | | Descriptor: | PROTEIN (NERVE GROWTH FACTOR RECEPTOR TRKA) | | Authors: | Wiesmann, C, Ultsch, M.H, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2RHK
 
 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | Authors: | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2SAS
 
 | |
1W83
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-CHLORO-3-(PYRIDIN-3-YLOXYMETHYL)-PHENYL]-3-FLUORO- | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
1WBT
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-5-MORPHOLIN-4-YL-N-[1-(2-PYRIDIN-4-YLETHYL)-1H-INDOL-6-YL]BENZAMIDE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
2WU8
 
 | |
1ZTS
 
 | | Solution Structure of Bacillus Subtilis Protein YQBG: Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Shen, Y, Xiao, R, Acton, T, Ma, L, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein yqbG from Bacillus subtilis reveals a novel alpha-helical protein fold.
Proteins, 62, 2006
|
|
2XA7
 
 | | AP2 clathrin adaptor core in active complex with cargo peptides | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, ALPHA 2 SUBUNIT, AP-2 COMPLEX SUBUNIT BETA, ... | | Authors: | Jackson, L.P, Kelly, B.T, McCoy, A.J, Evans, P.R, Owen, D.J. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Large Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the Ap2 Clathrin Adaptor Complex
Cell(Cambridge,Mass.), 141, 2010
|
|
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6H6K
 
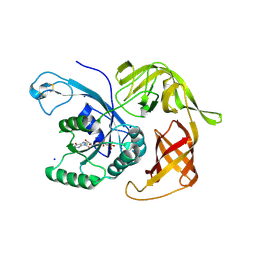 | | The structure of the FKR mutant of the archaeal translation initiation factor 2 gamma subunit in complex with GDPCP, obtained in the absence of magnesium salts in the crystallization solution. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, SODIUM ION, ... | | Authors: | Nikonov, O, Kravchenko, O, Nevskaya, N, Stolboushkina, E, Gabdulkhakov, A, Garber, M, Nikonov, S. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4MKW
 
 | |
4MRB
 
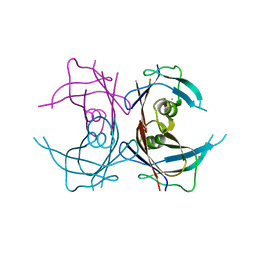 | | Wild Type Human Transthyretin pH 7.5 | | Descriptor: | CALCIUM ION, Transthyretin | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O1T
 
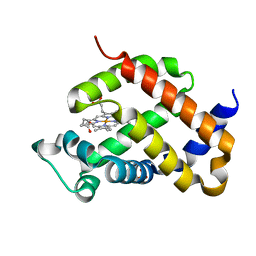 | | Crystal structure of murine neuroglobin mutant F106W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-16 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O1P
 
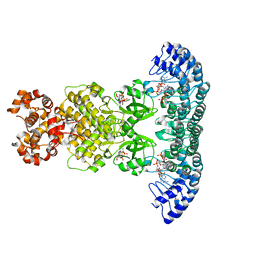 | | Crystal Structure of RNase L in complex with 2-5A and AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonuclease L, ... | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4NCG
 
 | | Discovery of Doravirine, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of MK-1439, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4O1O
 
 | | Crystal Structure of RNase L in complex with 2-5A | | Descriptor: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4MSL
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF40431 | | Descriptor: | N-[(7-hydroxy-4-methyl-2-oxo-2H-chromen-8-yl)methyl]-L-leucine, Sortilin, TETRAETHYLENE GLYCOL, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the first small-molecule ligand of the neuronal receptor sortilin and structure determination of the receptor-ligand complex.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P27
 
 | | Structure of Schistosoma mansoni venom allergen-like protein 4 (SmVAL4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Venom allergen-like (VAL) 4 protein | | Authors: | Asojo, O.A. | | Deposit date: | 2014-03-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Schistosoma mansoni venom allergen-like protein 4 (SmVAL4) is a novel lipid-binding SCP/TAPS protein that lacks the prototypical CAP motifs.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N58
 
 | | Crystal Structure of Pectocin M2 at 1.86 Angstroms | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Grinter, R, Roszak, A.W, Zeth, K, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
