6KSF
 
 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | Authors: | Li, H, Zhang, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
5DES
 
 | | 2009 H1N1 PA endonuclease domain | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5E83
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
5EP2
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain in Complex with AzaU Inhibitor | | Descriptor: | 2,2-dimethylpropyl 2-[[3,5-bis(oxidanylidene)-2~{H}-1,2,4-triazin-6-yl]sulfanyl]ethanoate, ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP1
 
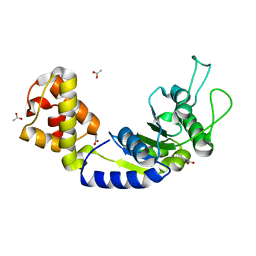 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain | | Descriptor: | ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP4
 
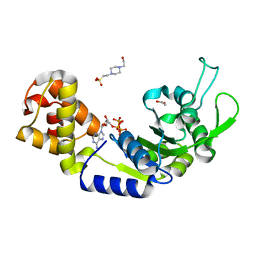 | | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
8JRN
 
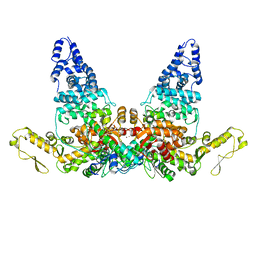 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRQ
 
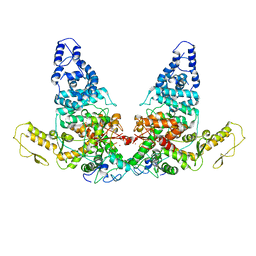 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRP
 
 | | Structure of E6AP-E6 complex in Att3 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRR
 
 | | Structure of E6AP-E6 complex in Det2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRO
 
 | | Structure of E6AP-E6 complex in Att2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
5EXH
 
 | |
5EP3
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain Bound to CV-133 Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,2-dimethylpropyl 2-[(3-oxidanylidene-5-sulfanylidene-2~{H}-1,2,4-triazin-6-yl)amino]ethanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5CZN
 
 | | 2009 H1N1 PA endonuclease mutant E119D | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8Z3K
 
 | | The structure of type III CRISPR-associated deaminase in complex 2cA6-2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
7VOK
 
 | |
8Z40
 
 | |
5EP0
 
 | | Quorum-Sensing Signal Integrator LuxO - Receiver+Catalytic Domains | | Descriptor: | 1,2-ETHANEDIOL, Putative repressor protein luxO, SULFATE ION | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
8Z3R
 
 | | The structure of type III CRISPR-associated deaminase in complex cA4 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Adenosine deaminase domain-containing protein, ZINC ION | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z3P
 
 | | The structure of type III CRISPR-associated deaminase in complex cA6 and ATP, fully activated | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
5DBS
 
 | | 2009 H1N1 PA endonuclease mutant E119D in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-21 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4OFZ
 
 | | Structure of unliganded trehalose-6-phosphate phosphatase from Brugia malayi | | Descriptor: | MAGNESIUM ION, Trehalose-phosphatase | | Authors: | Farelli, J.D, Allen, K.N, Carlow, C.K.S, Dunaway-Mariano, D. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Trehalose-6-phosphate Phosphatase from Brugia malayi Reveals Key Design Principles for Anthelmintic Drugs.
Plos Pathog., 10, 2014
|
|
4OAS
 
 | | co-crystal structure of MDM2 (17-111) in complex with compound 25 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-06 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AMG 232, a Potent, Selective, and Orally Bioavailable MDM2-p53 Inhibitor in Clinical Development.
J.Med.Chem., 57, 2014
|
|
3N9M
 
 | | ceKDM7A from C.elegans, alone | | Descriptor: | FE (II) ION, Putative uncharacterized protein, ZINC ION | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
4MD6
 
 | | Crystal structure of PDE5 in complex with inhibitor 5R | | Descriptor: | 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cui, W, Huang, M, Shao, Y, Luo, H. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one as a phosphodiesterase-5 inhibitor and its complex crystal structure.
Biochem Pharmacol, 89, 2014
|
|
