7WQB
 
 | | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132
To Be Published
|
|
7WQK
 
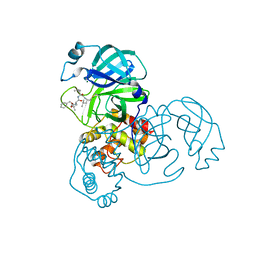 | | wild-type SARS-CoV-2 main protease in complex with MG-132 | | Descriptor: | 3C-like proteinase, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | wild-type SARS-CoV-2 main protease in complex with MG-132
To Be Published
|
|
7DNJ
 
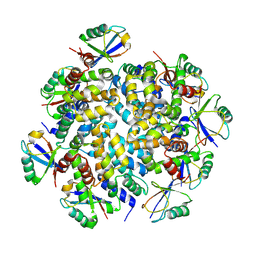 | | K63-polyUb MDA5CARDs complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7DNI
 
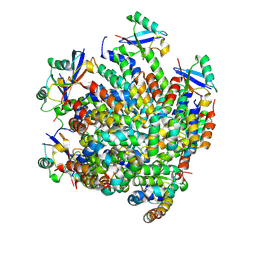 | | MDA5 CARDs-MAVS CARD polyUb complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Mitochondrial antiviral-signaling protein, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7BRE
 
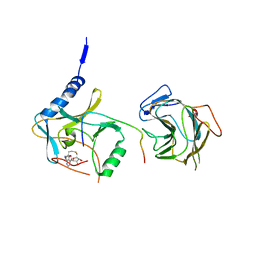 | | The crystal structure of MLL2 in complex with ASH2L and RBBP5 | | Descriptor: | Histone-lysine N-methyltransferase 2B, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Zhao, L, Chen, Y. | | Deposit date: | 2020-03-28 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal Structure of MLL2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by KMT2 Family Methyltransferases.
Structure, 28, 2020
|
|
7BQW
 
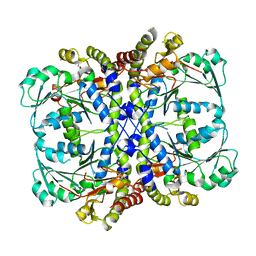 | |
7DTY
 
 | | Structural basis of ligand selectivity conferred by the human glucose-dependent insulinotropic polypeptide receptor | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhang, C, Zhou, Q.T, Hang, K.N, Zou, X.Y, Chen, Y, Wu, F, Rao, Q.D, Dai, A.T, Yin, W.C, Shen, D.D, Zhang, Y, Xia, T, Stevens, R.C, Xu, H.E, Yang, D.H, Zhao, L.H, Wang, M.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor.
Elife, 10, 2021
|
|
7CH1
 
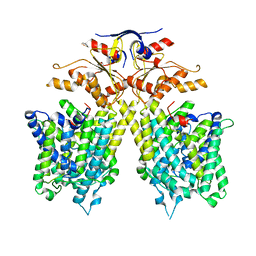 | | The overall structure of SLC26A9 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 26 member 9 | | Authors: | Chi, X.M, Chen, Y, Li, X.R, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence.
Cell Discov, 6, 2020
|
|
7F4D
 
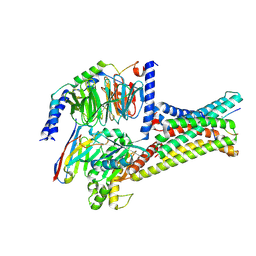 | | Cryo-EM structure of alpha-MSH-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4F
 
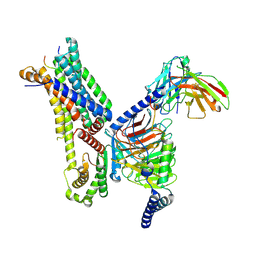 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4I
 
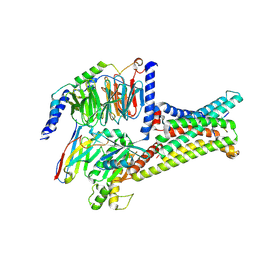 | | Cryo-EM structure of SHU9119-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4H
 
 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein, Nb35 and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
3NXH
 
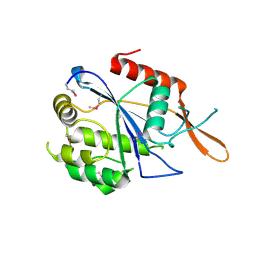 | | Crystal Structure of the transcriptional regulator yvhJ from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR735. | | Descriptor: | transcriptional regulator yvhJ | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-13 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Crystal Structure of the transcriptional regulator yvhJ from Bacillus subtilis.
To be Published
|
|
3P9V
 
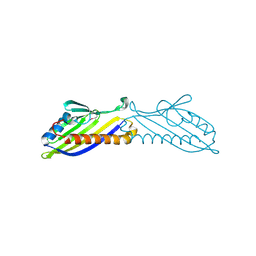 | | High Resolution Crystal Structure of protein Maqu_3174 from Marinobacter aquaeolei, Northeast Structural Genomics Consortium Target MqR197 | | Descriptor: | Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-10-18 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Northeast Structural Genomics Consortium Target MqR197
To be published
|
|
3PIL
 
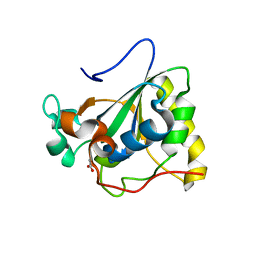 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | ACETATE ION, Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PIM
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PU2
 
 | | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides. Northeast Structural Genomics Consortium Target RhR263. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides.
To be Published
|
|
3PIN
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in complex with Trx2 | | Descriptor: | Peptide methionine sulfoxide reductase, Thioredoxin-2 | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3P51
 
 | | Three-dimensional structure of protein Q2Y8N9_NITMU from nitrosospira multiformis, Northeast structural genomics consortium target NMR118 | | Descriptor: | Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-10-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Northeast Structural Genomics Consortium Target NmR118
To be published
|
|
3QV0
 
 | | Crystal structure of Saccharomyces cerevisiae Mam33 | | Descriptor: | Mitochondrial acidic protein MAM33 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3P19
 
 | | Improved NADPH-dependent Blue Fluorescent Protein | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative blue fluorescent protein | | Authors: | Kao, T.H, Chen, Y, Pai, C.H, Wang, A.H.J. | | Deposit date: | 2010-09-30 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a NADPH-dependent blue fluorescent protein revealed the unique role of Gly176 on the fluorescence enhancement.
J.Struct.Biol., 174, 2011
|
|
3Q6A
 
 | | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116
To be Published
|
|
3QUW
 
 | | Crystal structure of yeast Mmf1 | | Descriptor: | Protein MMF1 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3RD6
 
 | | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403 | | Descriptor: | Mll3558 protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-01 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403
To be Published
|
|
3PUT
 
 | | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239. | | Descriptor: | HEXANE-1,6-DIOL, Hypothetical conserved protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239.
To be Published
|
|
