6LTQ
 
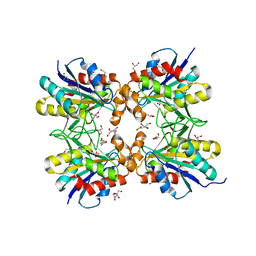 | | Crystal structure of pyrrolidone carboxyl peptidase from thermophilic keratin degrading bacterium Fervidobacterium islandicum AW-1 (FiPcp) | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Pyroglutamyl-peptidase I | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-01-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oxidized pyrrolidone carboxypeptidase from Fervidobacterium islandicum AW-1 reveals unique structural features for thermostability and keratinolysis.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
6FZU
 
 | |
6M2D
 
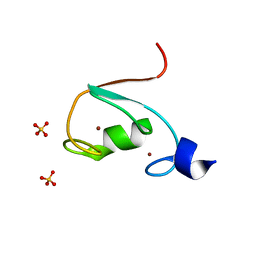 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
 | |
6G4T
 
 | |
8A6G
 
 | |
6G3C
 
 | | Crystal Structure of JAK2-V617F pseudokinase domain in complex with Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7,7-trimethyl-8-(3-methylbutyl)pteridin-6-one, Tyrosine-protein kinase | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
Acs Chem.Biol., 14, 2019
|
|
6G05
 
 | | RORGT (264-518;C455S) IN COMPLEX WITH INVERSE AGONIST "CPD-2" AND RIP140 PEPTIDE AT 1.90A | | Descriptor: | 2-(4-ethylsulfonylphenyl)-~{N}-[4-phenyl-5-(phenylcarbonyl)-1,3-thiazol-2-yl]ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimizing a Weakly Binding Fragment into a Potent ROR gamma t Inverse Agonist with Efficacy in an in Vivo Inflammation Model.
J. Med. Chem., 61, 2018
|
|
6N9G
 
 | | Crystal Structure of RGS7-Gbeta5 dimer | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Regulator of G-protein signaling 7 | | Authors: | Patil, D.N, Rangarajan, E, Izard, T, Martemyanov, K.A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Elife, 7, 2018
|
|
5FON
 
 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain (apo structure) | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
6G07
 
 | | RORGT (264-518;C455S) IN COMPLEX WITH INVERSE AGONIST "CPD-9" AND RIP140 PEPTIDE AT 1.66A | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1, ~{N}-[5-chloranyl-6-[(1~{S})-1-phenylethoxy]pyridin-3-yl]-2-(4-ethylsulfonylphenyl)ethanamide | | Authors: | Kallen, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Optimizing a Weakly Binding Fragment into a Potent ROR gamma t Inverse Agonist with Efficacy in an in Vivo Inflammation Model.
J. Med. Chem., 61, 2018
|
|
6Q9Z
 
 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
5FOM
 
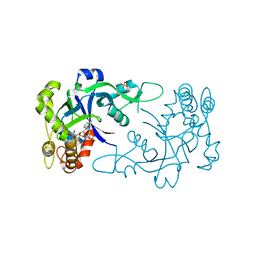 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with the adduct AMP-AN6426 | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, LEUCYL-TRNA SYNTHETASE, PHOSPHATE ION | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8A6R
 
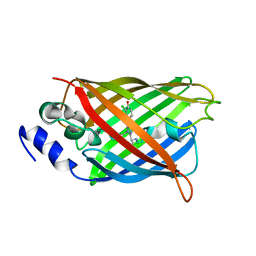 | |
8A6O
 
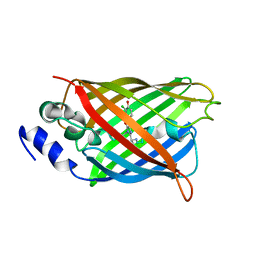 | |
8A6S
 
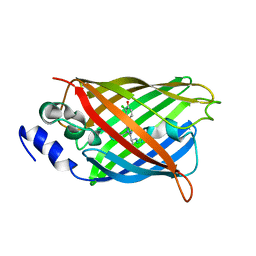 | |
8A6N
 
 | |
8A6P
 
 | |
8A6Q
 
 | |
8A7V
 
 | |
5FOL
 
 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of isoeucine (Ile2AA) | | Descriptor: | 2'-(L-ISOLEUCYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, PHOSPHATE ION | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8AM4
 
 | | Cl-rsEGFP2 Long Wavelength Structure | | Descriptor: | Green fluorescent protein | | Authors: | Orr, C.M, Fadini, A, van Thor, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Serial Femtosecond Crystallography Reveals that Photoactivation in a Fluorescent Protein Proceeds via the Hula Twist Mechanism.
J.Am.Chem.Soc., 2023
|
|
5LBR
 
 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at SACLA | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | Authors: | Edlund, P, Claesson, E, Nakane, T, Takala, H, Dods, R, Schmidt, M, Westenhoff, S. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
6NMV
 
 | | Non-Blocking Fab 218 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 218 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 218 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
