2MBS
 
 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
2M5N
 
 | | Atomic-resolution structure of a cross-beta protofilament | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-{beta} amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M7L
 
 | |
2MJE
 
 | | Reduced Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2M7X
 
 | | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2 | | Descriptor: | Na(+)/H(+) antiporter | | Authors: | Ullah, A, Kemp, G, Lee, B, Alves, C, Young, H, Sykes, B.D, Fliegel, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2.
J.Biol.Chem., 288, 2013
|
|
2LYS
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2M21
 
 | | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*UP*AP*CP*AP*CP*UP*AP*UP*UP*UP*AP*UP*CP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Wu, H, Trantirek, L, O'Connor, C.M, Feigon, J, Collins, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Rna, 12, 2006
|
|
2MBE
 
 | |
2MIJ
 
 | | NMR structure of the S-linked glycopeptide sublancin 168 | | Descriptor: | SPBc2 prophage-derived bacteriocin sublancin-168, beta-D-glucopyranose | | Authors: | Garcia De Gonzalo, C.V, Zhu, L, Oman, T.J, van der Donk, W.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the s-linked glycopeptide sublancin 168.
Acs Chem.Biol., 9, 2014
|
|
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
2MIP
 
 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS (HIV) TYPE 2 PROTEASE IN COMPLEX WITH A REDUCED AMIDE INHIBITOR AND COMPARISON WITH HIV-1 PROTEASE STRUCTURES | | Descriptor: | HIV-2 PROTEASE, INHIBITOR BI-LA-398 | | Authors: | Tong, L, Pav, S, Pargellis, C, Do, F, Lamarre, D, Anderson, P.C. | | Deposit date: | 1993-06-03 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human immunodeficiency virus (HIV) type 2 protease in complex with a reduced amide inhibitor and comparison with HIV-1 protease structures.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2MMS
 
 | | AG(7-deaza)G FAPY modified duplex | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*(7GU)P*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*CP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
2MKJ
 
 | | Solution structure of tandem RRM domains of cytoplasmic polyadenylation element binding protein 4 (CPEB4) in free state | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 4 | | Authors: | Afroz, T, Skrisovska, L, Belloc, E, Boixet, J.G, Mendez, R, Allain, F.H.-T. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins
Genes Dev., 28, 2014
|
|
2MHS
 
 | | NMR Structure of human Mcl-1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, G, Poppe, L, Aoki, K, Yamane, H, Lewis, J, Szyperski, T. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Quality NMR Structure of Human Anti-Apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design.
Plos One, 9, 2014
|
|
2M8L
 
 | | HIV capsid dimer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2MKM
 
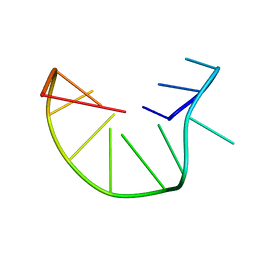 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3') | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2M00
 
 | |
2LYR
 
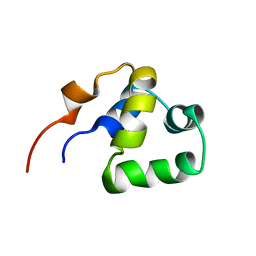 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2MKF
 
 | |
2MNA
 
 | | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | Single-stranded DNA binding protein (SSB), ssDNA | | Authors: | Gamsjaeger, R, Kariawasam, R, Gimenez, A.X, Touma, C.F, McIlwain, E, Bernardo, R.E, Shepherd, N.E, Ataide, S.F, Dong, A.Q, Richard, D.J, White, M.F, Cubeddu, L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus
Biochem.J., 465, 2015
|
|
2M1N
 
 | |
2M1E
 
 | |
4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4URE
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
7BET
 
 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
