8DG6
 
 | |
2MXP
 
 | |
6OSY
 
 | |
6OT1
 
 | |
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
7FAT
 
 | | Structure Determination of the RBD-NB1A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | Authors: | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
9II5
 
 | | Crystal structure of human TRIM21 PRYSPRY in complex with compound 1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ~{N}-[(1-fluoranylcyclohexyl)methyl]-~{N}-methyl-4-(2-methylsulfanylphenyl)-2-methylsulfonyl-benzamide | | Authors: | Zhang, L.Y. | | Deposit date: | 2024-06-19 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Chemically Induced Nuclear Pore Complex Protein Degradation via TRIM21.
Acs Chem.Biol., 20, 2025
|
|
2ATH
 
 | | Crystal structure of the ligand binding domain of human PPAR-gamma im complex with an agonist | | Descriptor: | 2-{5-[3-(7-PROPYL-3-TRIFLUOROMETHYLBENZO[D]ISOXAZOL-6-YLOXY)PROPOXY]INDOL-1-YL}ETHANOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Mahindroo, N, Huang, C.-F, Wu, S.-Y, Hsieh, H.-P. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Novel indole-based peroxisome proliferator-activated receptor agonists: design, SAR, structural biology, and biological activities
J.Med.Chem., 48, 2005
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
9B97
 
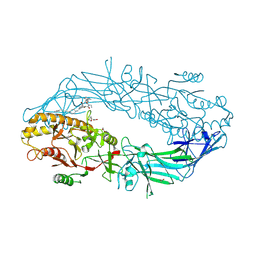 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (1P)-N~3'~-[(2S)-3-cyclohexyl-1-(methylamino)-1-oxopropan-2-yl]-N~3~,N~3~-diethyl-4-fluoro-5'-{[4-(4-phenylbutyl)piperazin-1-yl]methyl}[1,1'-biphenyl]-3,3'-dicarboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B96
 
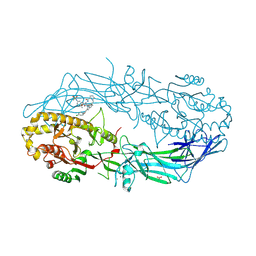 | | Crystal structure of the human PAD2 protein bound to inhibitor | | Descriptor: | 1-({(2P)-1-{(1R)-1-(2-bromophenyl)-3-[5-(methanesulfonamido)-2-methylanilino]-3-oxopropyl}-2-[3-(4-chlorophenoxy)phenyl]-1H-1,3-benzimidazol-6-yl}methyl)-N-methyl-D-prolinamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B98
 
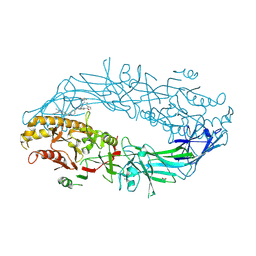 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (5P)-N,N-diethyl-2-fluoro-5-(2-[({1-[2-(methylamino)-2-oxoethyl]cyclohexyl}methyl)amino]-6-{methyl[1-(2-methyl-1-phenyl-1H-1,3-benzimidazole-5-carbonyl)piperidin-4-yl]amino}pyrimidin-4-yl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
5XGK
 
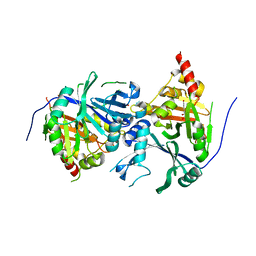 | | Crystal structure of Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase (AtHPPD) complexed with its substrate 4-hydroxyphenylpyruvate acid (HPPA) | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, 4-hydroxyphenylpyruvate dioxygenase, ACETATE ION, ... | | Authors: | Yang, G.F, Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-04-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Res, 2019, 2019
|
|
8H6T
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6P
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
6FC6
 
 | |
9JQR
 
 | |
9JQP
 
 | |
9JQ6
 
 | | Cryo-EM structure of BTN2A1 in complex with antagonist antibody TH002 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BTN2A1 antagonist antibody TH002-Fab heavy chain, ... | | Authors: | Zhang, M, Wang, Y.Q, Xiao, J.Y. | | Deposit date: | 2024-09-27 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of butyrophilin multimers reveal a plier-like mechanism for V gamma 9V delta 2 T cell receptor activation.
Immunity, 2025
|
|
9JQQ
 
 | | Cryo-EM structure of the HMBPP-primed BTN3A1-BTN3A2-BTN2A1 complex | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, M, Wang, Y.Q, Xiao, J.Y. | | Deposit date: | 2024-09-28 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of butyrophilin multimers reveal a plier-like mechanism for V gamma 9V delta 2 T cell receptor activation.
Immunity, 2025
|
|
