4WJZ
 
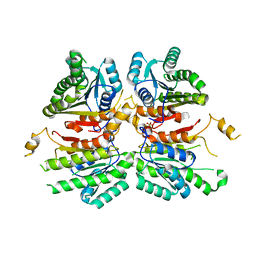 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(G141A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, PHOSPHATE ION | | Authors: | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4W8K
 
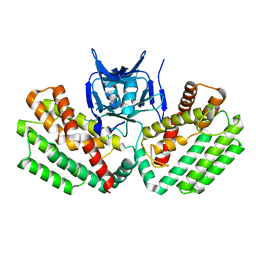 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
7TL5
 
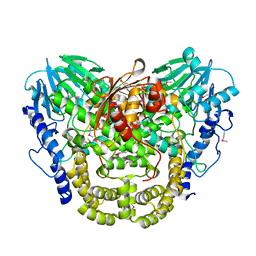 | | Crystal structure of putative hydrolase yjcS from Klebsiella pneumoniae. | | Descriptor: | 1,2-ETHANEDIOL, Lactamase_B domain-containing protein | | Authors: | Chang, C, Endres, M, Wu, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6W2Z
 
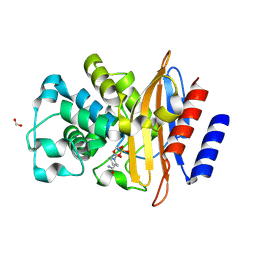 | | Crystal Structure of the Beta Lactamase Class A PenP from Bacillus subtilis in the Complex with the Non-beta- lactam Beta-lactamase Inhibitor Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-08 | | Release date: | 2020-03-25 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
6WN5
 
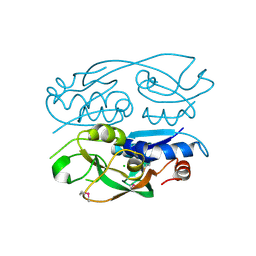 | | 1.52 Angstrom Resolution Crystal Structure of Transcriptional Regulator HdfR from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Transcriptional regulator HdfR | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WN8
 
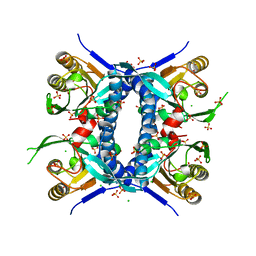 | | 2.70 Angstrom Resolution Crystal Structure of Uracil Phosphoribosyl Transferase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, SULFATE ION, Uracil phosphoribosyltransferase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
3LZM
 
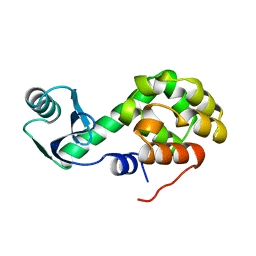 | |
6X1L
 
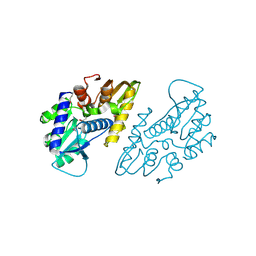 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
6PXA
 
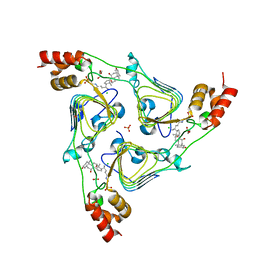 | | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid | | Descriptor: | ACETATE ION, CHLORIDE ION, Chloramphenicol acetyltransferase, ... | | Authors: | Tan, K, Maltseva, N, Jedrzejczak, R, Kuhn, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-25 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
5HM3
 
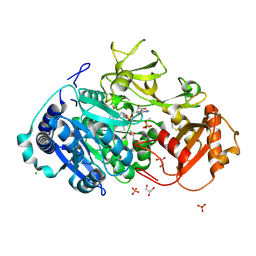 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
3IGS
 
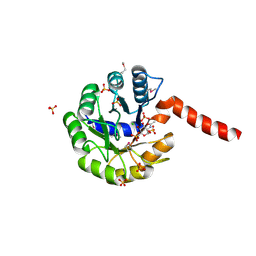 | | Structure of the Salmonella enterica N-acetylmannosamine-6-phosphate 2-epimerase | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, N-acetylmannosamine-6-phosphate 2-epimerase 2, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
3GOA
 
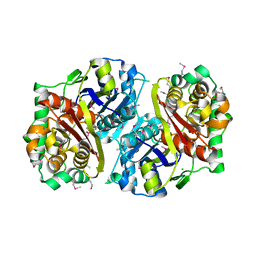 | | Crystal structure of the Salmonella typhimurium FadA 3-ketoacyl-CoA thiolase | | Descriptor: | 3-ketoacyl-CoA thiolase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, S.M, Skarina, T, Onopriyenko, O, Wawrzak, Z, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: |
|
|
7LA6
 
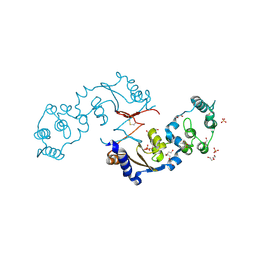 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239 deletion mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-05 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
7KB9
 
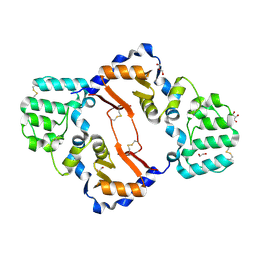 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, D238-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Sensor histidine kinase | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
6CTY
 
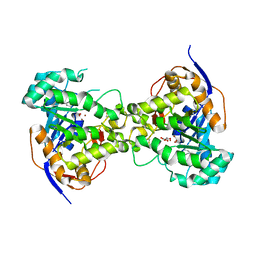 | | Crystal structure of dihydroorotase pyrC from Yersinia pestis in complex with zinc and malate at 2.4 A resolution | | Descriptor: | D-MALATE, Dihydroorotase, ZINC ION | | Authors: | Lipowska, J, Shabalin, I.G, Winsor, J, Woinska, M, Cooper, D.R, Kwon, K, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
4ZTH
 
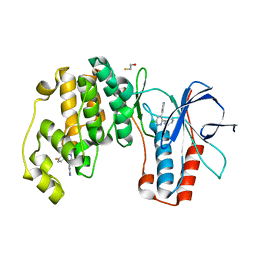 | | Structure of human p38aMAPK-arylpyridazinylpyridine fragment complex used in inhibitor discovery | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-chloro-3-phenyl-4-(pyridin-4-yl)pyridazine, BETA-MERCAPTOETHANOL, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Watterson, D.M. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
2R6O
 
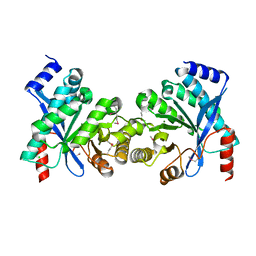 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Thiobacillus denitrificans | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
4JQO
 
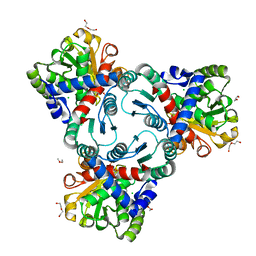 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with citrulline and inorganic phosphate | | Descriptor: | CHLORIDE ION, CITRULLINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Domagalski, M.J, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4JFR
 
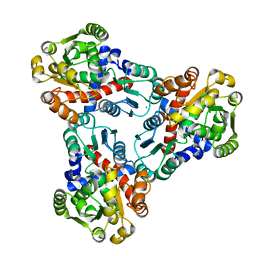 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Bajor, J, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
to be published
|
|
4JHX
 
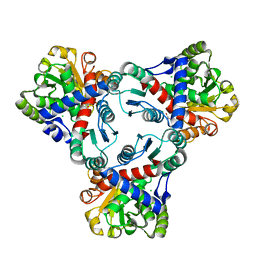 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoylphosphate and arginine | | Descriptor: | ARGININE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Bacal, P, Winsor, J, Grimshaw, S, Grabowski, M, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
1UP7
 
 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
