4R95
 
 | | BACE-1 in complex with 2-(((1R,3S)-3-(((R)-4-(2-cyclohexylethyl)-2-iminio-1-methyl-5-oxoimidazolidin-4-yl)methyl)cyclohexyl)amino)quinolin-1-ium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-2-imino-3-methyl-5-{[(1S,3R)-3-(quinolin-2-ylamino)cyclohexyl]methyl}imidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R8Y
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4O7X
 
 | |
4OK6
 
 | |
4OK3
 
 | |
4OKS
 
 | |
7DTL
 
 | | Crystal structure of PSK, an antimicrobial peptide from Chrysomya megacephala | | Descriptor: | PSK | | Authors: | Xiao, C, Xiao, Z, Wang, S, Liu, W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3TDA
 
 | | Competitive replacement of thioridazine by prinomastat in crystals of cytochrome P450 2D6 | | Descriptor: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, Prinomastat, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4O3O
 
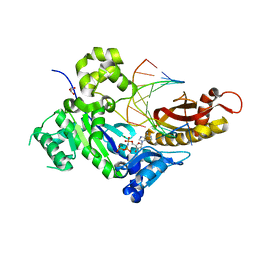 | | Crystal structure of human polymerase eta inserting datp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O62
 
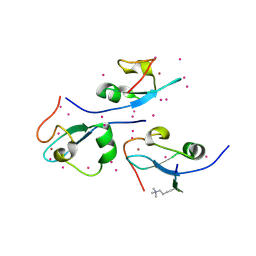 | | CW-type zinc finger of ZCWPW2 in complex with the amino terminus of histone H3 | | Descriptor: | Histone H3.3, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-26 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
4O3S
 
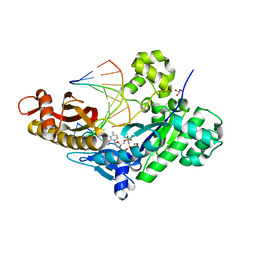 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-dc pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
7E5X
 
 | |
4O3P
 
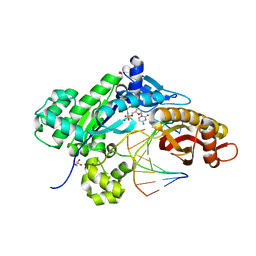 | | Crystal structure of human polymerase eta inserting dctp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4NRO
 
 | |
4O3Q
 
 | | Crystal structure of human polymerase eta inserting dgtp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4NRM
 
 | |
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp4/5 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp14/15 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3TBG
 
 | | Human cytochrome P450 2D6 with two thioridazines bound in active site | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Cytochrome P450 2D6, GLYCEROL, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4QEC
 
 | | ElxO with NADP Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
