2RJ8
 
 | |
2RJ1
 
 | |
3I6E
 
 | | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM Ruegeria pomeroyi. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase I, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of muconate lactonizing enzyme from Ruegeria pomeroyi.
To be Published
|
|
3IJI
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; nonproductive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IK4
 
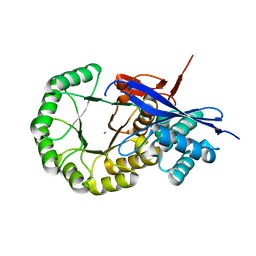 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2RJ6
 
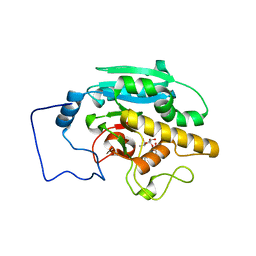 | |
2RJ5
 
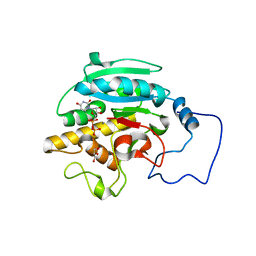 | |
3I7Q
 
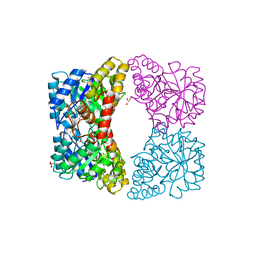 | | Dihydrodipicolinate synthase mutant - K161A | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
2RIZ
 
 | |
3IFV
 
 | | Crystal structure of the Haloferax volcanii proliferating cell nuclear antigen | | Descriptor: | PCNA, SODIUM ION | | Authors: | Winter, J.A, Christofi, P, Morroll, S, Bunting, K.A. | | Deposit date: | 2009-07-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Haloferax volcanii proliferating cell nuclear antigen reveals unique surface charge characteristics due to halophilic adaptation
Bmc Struct.Biol., 9, 2009
|
|
3IJQ
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1DWV
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
3ID8
 
 | | Ternary complex of human pancreatic glucokinase crystallized with activator, glucose and AMP-PNP | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, Glucokinase, MAGNESIUM ION, ... | | Authors: | Petit, P, Gluais, L, Lagarde, A, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2009-07-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3IFB
 
 | | NMR STUDY OF HUMAN INTESTINAL FATTY ACID BINDING PROTEIN | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein: implications for ligand entry and exit.
J.Biomol.NMR, 9, 1997
|
|
3IFX
 
 | | Crystal structure of the Spin-labeled KcsA mutant V48R1 | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, TETRABUTYLAMMONIUM ION, ... | | Authors: | Cieslak, J.A, Focia, P.J, Gross, A. | | Deposit date: | 2009-07-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | EPR (3.56 Å), X-RAY DIFFRACTION | | Cite: | Electron Spin-Echo Envelope Modulation (ESEEM) Reveals Water and Phosphate Interactions with the KcsA Potassium Channel
Biochemistry, 49, 2010
|
|
1DWX
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
3IWE
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646 | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646
To be Published
|
|
3IJL
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Pro-D-Glu; nonproductive substrate binding. | | Descriptor: | D-GLUTAMIC ACID, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7SO6
 
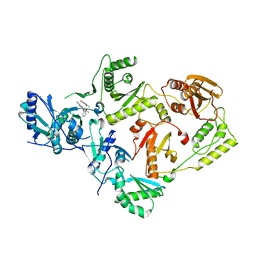 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO4
 
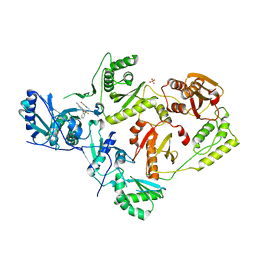 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
3I7R
 
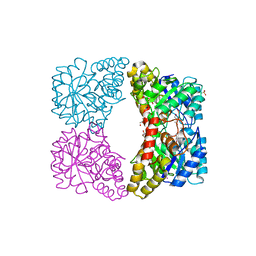 | | Dihydrodipicolinate synthase - K161R | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
3IU1
 
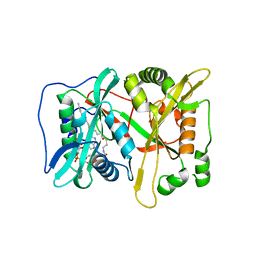 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA
To be Published
|
|
3I7S
 
 | | Dihydrodipicolinate synthase mutant - K161A - with the substrate pyruvate bound in the active site. | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
3IK2
 
 | | Crystal Structure of a Glycoside Hydrolase Family 44 Endoglucanase produced by Clostridium acetobutylium ATCC 824 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Warner, C.D, Hoy, J.A, Ford, C.F, Honzatko, R.B, Reilly, P.J. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tertiary structure and characterization of a glycoside hydrolase family 44 endoglucanase from Clostridium acetobutylicum.
Appl.Environ.Microbiol., 76, 2010
|
|
3IDH
 
 | | Human pancreatic glucokinase in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, alpha-D-glucopyranose | | Authors: | Petit, P, Gluais, L, Lagarde, A, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2009-07-21 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
