6TRK
 
 | | Phl p 6 fold stabilized mutant - S46Y | | Descriptor: | Pollen allergen Phl p 6, ZINC ION | | Authors: | Soh, W.T, Brandstetter, H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In silico Design of Phl p 6 Variants With Altered Fold-Stability Significantly Impacts Antigen Processing, Immunogenicity and Immune Polarization.
Front Immunol, 11, 2020
|
|
8US8
 
 | |
4RX0
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
1VTQ
 
 | | THREE-DIMENSIONAL STRUCTURE OF YEAST T-RNA-ASP. I. STRUCTURE DETERMINATION | | Descriptor: | T-RNA-ASP | | Authors: | Comarmond, M.B, Giege, R, Thierry, J.C, Moras, D, Fischer, J. | | Deposit date: | 1985-06-11 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of Yeast T-RNA-ASP. I. Structure Determination
Acta Crystallogr.,Sect.B, 42, 1986
|
|
1MZI
 
 | | Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope | | Descriptor: | 2F5 epitope of HIV-1 gp41 fusion protein | | Authors: | Barbato, G, Bianchi, E, Ingallinella, P, Hurni, W.H, Miller, M.D, Ciliberto, G, Cortese, R, Bazzo, R, Shiver, J.W, Pessi, A. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion.
J.Mol.Biol., 330, 2003
|
|
7A9C
 
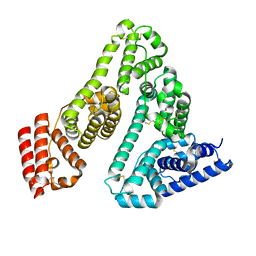 | |
6T41
 
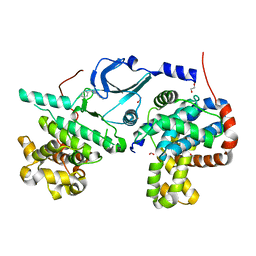 | | CDK8/Cyclin C in complex with N-(4-chlorobenzyl)isoquinolin-4-amine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Maskos, K, Huber, R, Kuhn, C.-D. | | Deposit date: | 2019-10-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A precisely positioned MED12 activation helix stimulates CDK8 kinase activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5J9A
 
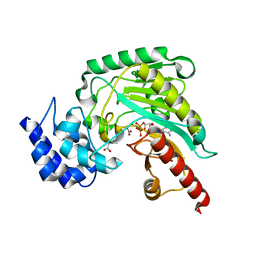 | | Ambient temperature transition state structure of arginine kinase - crystal 11/Form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
8FBT
 
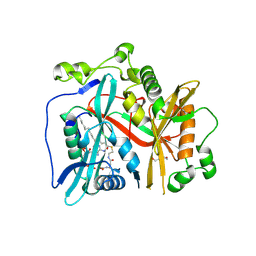 | | Crystal structure of Cryptosporidium parvum N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Staker, B.L, Fenwick, M.K, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of and Structural Insights into Hit Compounds Targeting N -Myristoyltransferase for Cryptosporidium Drug Development.
Acs Infect Dis., 9, 2023
|
|
8FBM
 
 | |
8FBU
 
 | | Crystal structure of Cryptosporidium parvum N-myristoyltransferase with bound myristoyl-CoA and Compound-2 | | Descriptor: | (2M)-2-[2-(piperazin-1-yl)phenyl]-N-(1,3-thiazol-2-yl)-1H-benzimidazole-4-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Staker, B.L, Fenwick, M.K, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of and Structural Insights into Hit Compounds Targeting N -Myristoyltransferase for Cryptosporidium Drug Development.
Acs Infect Dis., 9, 2023
|
|
4Z3L
 
 | |
8FBQ
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 12b | | Descriptor: | 1-[(3M)-3-{3-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]pyridin-2-yl}phenyl]piperazine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Staker, B.L, Lovell, S.W, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of potent and selective N-myristoyltransferase inhibitors of Plasmodium vivax liver stage hypnozoites and schizonts.
Nat Commun, 14, 2023
|
|
8F82
 
 | |
8F84
 
 | |
8F80
 
 | |
8F83
 
 | |
8F85
 
 | |
8F81
 
 | |
7Z94
 
 | | Crystal structure of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with indole | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, INDOLE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Z4X
 
 | |
7Z98
 
 | | Crystal structure of F191M variant Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with methyl phenyl sulfide | | Descriptor: | (methylsulfanyl)benzene, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7ZCA
 
 | | Crystal structure of the F191M/F201A variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with benzyl phenyl sulfoxide | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative dehydrogenase/oxygenase subunit (Flavoprotein), ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-26 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Z99
 
 | |
7Z97
 
 | | Crystal structure of the F191M variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with 6-bromoindole | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
