3TH9
 
 | | Crystal Structure of HIV-1 Protease Mutant Q7K V32I L63I with a cyclic sulfonamide inhibitor | | Descriptor: | Gag-Pol polyprotein, tert-butyl {(2S,3R)-4-[(4S)-7-fluoro-4-methyl-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate | | Authors: | Orth, P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design, Synthesis, and X-ray Crystallographic Analysis of a Novel Class of HIV-1 Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
8IO9
 
 | |
8IOA
 
 | |
8IO8
 
 | |
8IO7
 
 | |
8IO6
 
 | |
8IOE
 
 | |
8XID
 
 | |
8K3Y
 
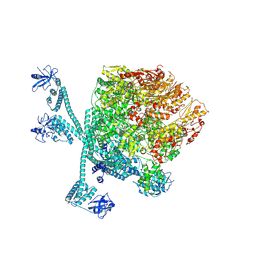 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
4MC9
 
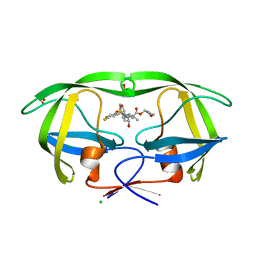 | | HIV protease in complex with AA74 | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
7W6M
 
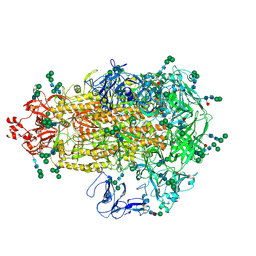 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W73
 
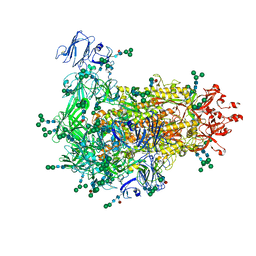 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-08-03 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6T
 
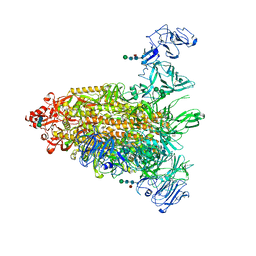 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6U
 
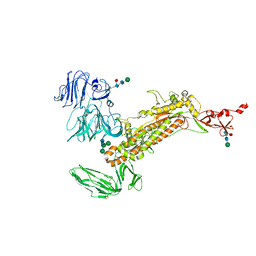 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6V
 
 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6S
 
 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
4OM5
 
 | | Crystal structure of CTX A4 from Taiwan Cobra (Naja naja atra) | | Descriptor: | Cytotoxin 4 | | Authors: | Lin, C.C, Chang, C.I, Wu, W.G. | | Deposit date: | 2014-01-26 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface.
J.Biol.Chem., 289, 2014
|
|
4OM4
 
 | | Crystal structure of CTX A2 from Taiwan Cobra (Naja naja atra) | | Descriptor: | Cytotoxin 2 | | Authors: | Lin, C.C, Chang, C.I, Wu, W.G. | | Deposit date: | 2014-01-26 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface.
J.Biol.Chem., 289, 2014
|
|
6LUS
 
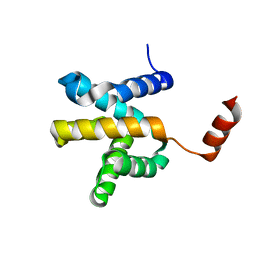 | | Crystal structure of the Mengla Virus VP30 C-terminal domain | | Descriptor: | Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Wen, K.N, Chu, H.G, Wang, C.H, Qin, X.C. | | Deposit date: | 2020-01-30 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Mengla virus VP30 C-terminal domain.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
8JBQ
 
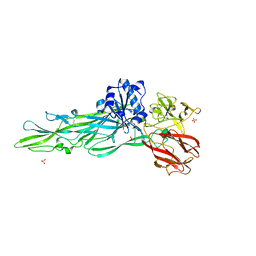 | |
7WCV
 
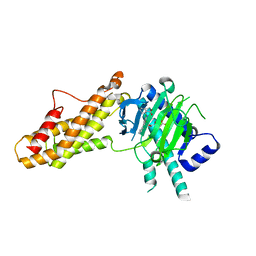 | | Co-crystal structure of FTO bound to 6e | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-pyridin-4-yl-phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationships and Antileukemia Effects of the Tricyclic Benzoic Acid FTO Inhibitors.
J.Med.Chem., 65, 2022
|
|
7XPJ
 
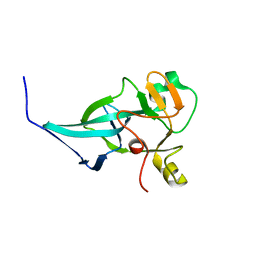 | | crystal structure of rice ASI1 BAH domain | | Descriptor: | BAH domain-containing protein | | Authors: | Yuan, J, Du, J. | | Deposit date: | 2022-05-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Molecular basis of locus-specific H3K9 methylation catalyzed by SUVH6 in plants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XPK
 
 | |
